TALEN-mediated enhancer knockout influences TNFAIP3 gene expression and mimics a molecular phenotype associated with systemic lupus erythematosus
- PMID: 26821284
- PMCID: PMC4840072
- DOI: 10.1038/gene.2016.4
TALEN-mediated enhancer knockout influences TNFAIP3 gene expression and mimics a molecular phenotype associated with systemic lupus erythematosus
Abstract
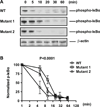
Linkage disequilibrium poses a major challenge to the functional characterization of specific disease-associated susceptibility variants. Precision genome-editing technologies have provided new opportunities to address this challenge. As proof of concept, we employed TALEN (transcription activation-like effector nuclease)-mediated genome editing to specifically disrupt the TT>A enhancer region to mimic candidate causal variants identified in the systemic lupus erythematosus-associated susceptibility gene, tumor necrosis factor-α-induced protein 3 (TNFAIP3), in an isogenic HEK293T cell line devoid of other linkage disequilibrium-associated variants. Targeted disruption of the TT>A enhancer impaired its interaction with the TNFAIP3 promoter by long-range DNA looping, thereby reducing TNFAIP3 gene expression. Loss of TNFAIP3 mRNA and its encoded protein, A20, impaired tumor necrosis factor-α-induced receptor-mediated downregulation of nuclear factor-κB signaling, a hallmark of autoimmunity. Results demonstrate that the TT>A enhancer variants contribute to causality and function independently of other variants to disrupt TNFAIP3 expression. Furthermore, we believe this approach can be implemented to independently examine other candidate casual variants in the future.
Conflict of interest statement
Figures

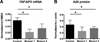


Similar articles
-
An enhancer element harboring variants associated with systemic lupus erythematosus engages the TNFAIP3 promoter to influence A20 expression.PLoS Genet. 2013;9(9):e1003750. doi: 10.1371/journal.pgen.1003750. Epub 2013 Sep 5. PLoS Genet. 2013. PMID: 24039598 Free PMC article.
-
Functional variants of TNFAIP3 are associated with systemic lupus erythematosus in a cohort of Chinese Han population.Hum Immunol. 2019 Feb;80(2):140-145. doi: 10.1016/j.humimm.2018.11.008. Epub 2018 Dec 5. Hum Immunol. 2019. PMID: 30529365
-
Synergistic activation of NF-κB by TNFAIP3 (A20) reduction and UBE2L3 (UBCH7) augment that synergistically elevate lupus risk.Arthritis Res Ther. 2020 Apr 25;22(1):93. doi: 10.1186/s13075-020-02181-4. Arthritis Res Ther. 2020. PMID: 32334614 Free PMC article.
-
A20/Tumor Necrosis Factor α-Induced Protein 3 in Immune Cells Controls Development of Autoinflammation and Autoimmunity: Lessons from Mouse Models.Front Immunol. 2018 Feb 21;9:104. doi: 10.3389/fimmu.2018.00104. eCollection 2018. Front Immunol. 2018. PMID: 29515565 Free PMC article. Review.
-
Roles of A20 in autoimmune diseases.Immunol Res. 2016 Apr;64(2):337-44. doi: 10.1007/s12026-015-8677-6. Immunol Res. 2016. PMID: 26135958 Review.
Cited by
-
Prioritizing disease and trait causal variants at the TNFAIP3 locus using functional and genomic features.Nat Commun. 2020 Mar 6;11(1):1237. doi: 10.1038/s41467-020-15022-4. Nat Commun. 2020. PMID: 32144282 Free PMC article.
-
The landscape of GWAS validation; systematic review identifying 309 validated non-coding variants across 130 human diseases.BMC Med Genomics. 2022 Apr 1;15(1):74. doi: 10.1186/s12920-022-01216-w. BMC Med Genomics. 2022. PMID: 35365203 Free PMC article.
-
A20/TNFAIP3 heterozygosity predisposes to behavioral symptoms in a mouse model for neuropsychiatric lupus.Brain Behav Immun Health. 2019 Dec 14;2:100018. doi: 10.1016/j.bbih.2019.100018. eCollection 2020 Feb. Brain Behav Immun Health. 2019. PMID: 38377433 Free PMC article.
-
An Autoimmune Disease-Associated Risk Variant in the TNFAIP3 Gene Plays a Protective Role in Brucellosis That Is Mediated by the NF-κB Signaling Pathway.J Clin Microbiol. 2018 Mar 26;56(4):e01363-17. doi: 10.1128/JCM.01363-17. Print 2018 Apr. J Clin Microbiol. 2018. PMID: 29343543 Free PMC article.
-
Hypomorphic A20 expression confers susceptibility to psoriasis.PLoS One. 2017 Jun 28;12(6):e0180481. doi: 10.1371/journal.pone.0180481. eCollection 2017. PLoS One. 2017. PMID: 28658319 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

