Squamous Cell Carcinoma-related Oncogene (SCCRO) Family Members Regulate Cell Growth and Proliferation through Their Cooperative and Antagonistic Effects on Cullin Neddylation
- PMID: 26792857
- PMCID: PMC4813546
- DOI: 10.1074/jbc.M115.692756
Squamous Cell Carcinoma-related Oncogene (SCCRO) Family Members Regulate Cell Growth and Proliferation through Their Cooperative and Antagonistic Effects on Cullin Neddylation
Abstract
SCCRO (squamous cell carcinoma-related oncogene; also known as DCUN1D1) is a highly conserved gene that functions as an E3 in neddylation. Although inactivation of SCCRO in yeast results in lethality, SCCRO(-/-) mice are viable. The exclusive presence of highly conserved paralogues in higher organisms led us to assess whether compensation by SCCRO paralogues rescues lethality in SCCRO(-/-) mice. Using murine and Drosophila models, we assessed the in vivo activities of SCCRO and its paralogues in cullin neddylation. We found that SCCRO family members have overlapping and antagonistic activity that regulates neddylation and cell proliferation activities in vivo. In flies, both dSCCRO and dSCCRO3 promote neddylation and cell proliferation, whereas dSCCRO4 negatively regulates these processes. Analysis of somatic clones showed that the effects that these paralogues have on proliferation serve to promote cell competition, leading to apoptosis in clones with a net decrease in neddylation activity. We found that dSCCRO and, to a lesser extent, dSCCRO3 rescue the neddylation and proliferation defects promoted by expression of SCCRO4. dSCCRO and dSCCRO3 functioned cooperatively, with their coexpression resulting in an increase in both the neddylated cullin fraction and proliferation activity. In contrast, human SCCRO and SCCRO4 promote, and human SCCRO3 inhibits, neddylation and proliferation when expressed in flies. Our findings provide the first insights into the mechanisms through which SCCRO family members cooperatively regulate neddylation and cell proliferation.
Keywords: head and neck cancer; lung cancer; oncogene; tumor suppressor gene; ubiquitylation (ubiquitination).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



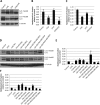
Similar articles
-
Squamous cell carcinoma-related oncogene (SCCRO) neddylates Cul3 protein to selectively promote midbody localization and activity of Cul3KLHL21 protein complex during abscission.J Biol Chem. 2017 Sep 15;292(37):15254-15265. doi: 10.1074/jbc.M117.778530. Epub 2017 Jun 15. J Biol Chem. 2017. PMID: 28620047 Free PMC article.
-
SCCRO3 (DCUN1D3) antagonizes the neddylation and oncogenic activity of SCCRO (DCUN1D1).J Biol Chem. 2014 Dec 12;289(50):34728-42. doi: 10.1074/jbc.M114.585505. Epub 2014 Oct 27. J Biol Chem. 2014. PMID: 25349211 Free PMC article.
-
SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation.J Biol Chem. 2008 Nov 28;283(48):33211-20. doi: 10.1074/jbc.M804440200. Epub 2008 Sep 30. J Biol Chem. 2008. PMID: 18826954 Free PMC article.
-
DCUN1D1 and neddylation: Potential targets for cancer therapy.Biochim Biophys Acta Mol Basis Dis. 2024 Oct;1870(7):167308. doi: 10.1016/j.bbadis.2024.167308. Epub 2024 Jun 15. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 38885797 Review.
-
Function and regulation of protein neddylation. 'Protein modifications: beyond the usual suspects' review series.EMBO Rep. 2008 Oct;9(10):969-76. doi: 10.1038/embor.2008.183. Epub 2008 Sep 19. EMBO Rep. 2008. PMID: 18802447 Free PMC article. Review.
Cited by
-
Emerging mechanisms of cell competition.Nat Rev Genet. 2020 Nov;21(11):683-697. doi: 10.1038/s41576-020-0262-8. Epub 2020 Aug 10. Nat Rev Genet. 2020. PMID: 32778819 Free PMC article. Review.
-
Improvement of Oral Bioavailability of Pyrazolo-Pyridone Inhibitors of the Interaction of DCN1/2 and UBE2M.J Med Chem. 2021 May 13;64(9):5850-5862. doi: 10.1021/acs.jmedchem.1c00035. Epub 2021 May 4. J Med Chem. 2021. PMID: 33945681 Free PMC article.
-
Mouse DCUN1D1 (SCCRO) is required for spermatogenetic individualization.PLoS One. 2019 Jan 17;14(1):e0209995. doi: 10.1371/journal.pone.0209995. eCollection 2019. PLoS One. 2019. PMID: 30653527 Free PMC article.
-
Squamous cell carcinoma-related oncogene (SCCRO) neddylates Cul3 protein to selectively promote midbody localization and activity of Cul3KLHL21 protein complex during abscission.J Biol Chem. 2017 Sep 15;292(37):15254-15265. doi: 10.1074/jbc.M117.778530. Epub 2017 Jun 15. J Biol Chem. 2017. PMID: 28620047 Free PMC article.
-
Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.Nat Chem Biol. 2017 Aug;13(8):850-857. doi: 10.1038/nchembio.2386. Epub 2017 Jun 5. Nat Chem Biol. 2017. PMID: 28581483 Free PMC article.
References
-
- Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Finley D., and Chau V. (1991) Ubiquitination. Annu. Rev. Cell Biol. 7, 25–69 - PubMed
-
- Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 - PubMed
-
- Berndsen C. E., and Wolberger C. (2014) New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

