The Rag-Ragulator Complex Regulates Lysosome Function and Phagocytic Flux in Microglia
- PMID: 26774477
- PMCID: PMC4731305
- DOI: 10.1016/j.celrep.2015.12.055
The Rag-Ragulator Complex Regulates Lysosome Function and Phagocytic Flux in Microglia
Abstract
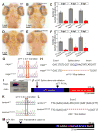
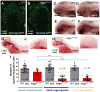
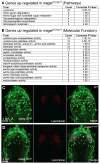
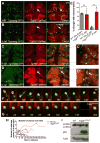
Microglia are resident macrophages of the CNS that are essential for phagocytosis of apoptotic neurons and weak synapses during development. We show that RagA and Lamtor4, two components of the Rag-Ragulator complex, are essential regulators of lysosomes in microglia. In zebrafish lacking RagA function, microglia exhibit an expanded lysosomal compartment, but they are unable to properly digest apoptotic neuronal debris. Previous biochemical studies have placed the Rag-Ragulator complex upstream of mTORC1 activation in response to cellular nutrient availability. Nonetheless, RagA and mTOR mutant zebrafish have distinct phenotypes, indicating that the Rag-Ragulator complex has functions independent of mTOR signaling. Our analysis reveals an essential role of the Rag-Ragulator complex in proper lysosome function and phagocytic flux in microglia.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Disruption of the Rag-Ragulator Complex by c17orf59 Inhibits mTORC1.Cell Rep. 2015 Sep 1;12(9):1445-55. doi: 10.1016/j.celrep.2015.07.052. Epub 2015 Aug 20. Cell Rep. 2015. PMID: 26299971 Free PMC article.
-
Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms.Proc Natl Acad Sci U S A. 2018 Sep 18;115(38):9545-9550. doi: 10.1073/pnas.1811727115. Epub 2018 Sep 4. Proc Natl Acad Sci U S A. 2018. PMID: 30181260 Free PMC article.
-
Rag-Ragulator is the central organizer of the physical architecture of the mTORC1 nutrient-sensing pathway.Proc Natl Acad Sci U S A. 2024 Aug 27;121(35):e2322755121. doi: 10.1073/pnas.2322755121. Epub 2024 Aug 20. Proc Natl Acad Sci U S A. 2024. PMID: 39163330
-
Rag GTPase in amino acid signaling.Amino Acids. 2016 Apr;48(4):915-928. doi: 10.1007/s00726-016-2171-x. Epub 2016 Jan 18. Amino Acids. 2016. PMID: 26781224 Review.
-
Amino acid signalling upstream of mTOR.Nat Rev Mol Cell Biol. 2013 Mar;14(3):133-9. doi: 10.1038/nrm3522. Epub 2013 Jan 30. Nat Rev Mol Cell Biol. 2013. PMID: 23361334 Free PMC article. Review.
Cited by
-
Microglial mTOR is Neuronal Protective and Antiepileptogenic in the Pilocarpine Model of Temporal Lobe Epilepsy.J Neurosci. 2020 Sep 30;40(40):7593-7608. doi: 10.1523/JNEUROSCI.2754-19.2020. Epub 2020 Aug 31. J Neurosci. 2020. PMID: 32868461 Free PMC article.
-
The Multifaceted Role of Nutrient Sensing and mTORC1 Signaling in Physiology and Aging.Front Aging. 2021 Aug 27;2:707372. doi: 10.3389/fragi.2021.707372. eCollection 2021. Front Aging. 2021. PMID: 35822019 Free PMC article. Review.
-
Deep post-GWAS analysis identifies potential risk genes and risk variants for Alzheimer's disease, providing new insights into its disease mechanisms.Sci Rep. 2021 Oct 15;11(1):20511. doi: 10.1038/s41598-021-99352-3. Sci Rep. 2021. PMID: 34654853 Free PMC article.
-
Hexb enzyme deficiency leads to lysosomal abnormalities in radial glia and microglia in zebrafish brain development.Glia. 2019 Sep;67(9):1705-1718. doi: 10.1002/glia.23641. Epub 2019 May 29. Glia. 2019. PMID: 31140649 Free PMC article.
-
Insights Into Central Nervous System Glial Cell Formation and Function From Zebrafish.Front Cell Dev Biol. 2021 Nov 29;9:754606. doi: 10.3389/fcell.2021.754606. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34912801 Free PMC article. Review.
References
-
- Appelqvist H, Wäster P, Kågedal K, Öllinger K. The lysosome: from waste bag to potential therapeutic target. Journal of Molecular Cell Biology. 2013;5(4):214–226. - PubMed
-
- Ballabio A, Gieselmann V. Lysosomal disorders: From storage to cellular damage. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2009;1793(4):684–696. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

