Chi and dLMO function antagonistically on Notch signaling through directly regulation of fng transcription
- PMID: 26738424
- PMCID: PMC4704065
- DOI: 10.1038/srep18937
Chi and dLMO function antagonistically on Notch signaling through directly regulation of fng transcription
Abstract
Gene apterous (ap), chip (chi) and beadex (bx) play important roles in the dorsal-ventral compartmentalization in Drosophila wing discs. Meanwhile, Notch signaling is essential to the same process. It has been reported that Ap and Chi function as a tetramer to regulate Notch signaling. At the same time, dLMO (the protein product of gene bx) regulates the activity of Ap by competing its binding with Chi. However, the detailed functions of Chi and dLMO on Notch signaling and the relevant mechanisms remain largely unknown. Here, we report the detailed functions of Chi and dLMO on Notch signaling. Different Chi protein levels in adjacent cells could activate Notch signaling mainly in the cells with higher level of Chi. dLMO could induce antagonistical phenotypes on Notch signaling compared to that induced by Chi. These processes depend on their direct regulation of fringe (fng) transcription.
Figures
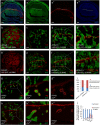
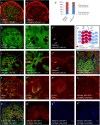
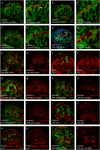
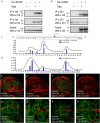
Similar articles
-
Fringe-dependent separation of dorsal and ventral cells in the Drosophila wing.Nature. 1999 Sep 30;401(6752):476-80. doi: 10.1038/46786. Nature. 1999. PMID: 10519550
-
The glycosyltransferase Fringe promotes Delta-Notch signaling between neurons and glia, and is required for subtype-specific glial gene expression.Development. 2007 Feb;134(3):591-600. doi: 10.1242/dev.02754. Development. 2007. PMID: 17215308
-
Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function.Genes Dev. 1998 Sep 15;12(18):2912-20. doi: 10.1101/gad.12.18.2912. Genes Dev. 1998. PMID: 9744867 Free PMC article.
-
Fringe: defining borders by regulating the notch pathway.Curr Opin Neurobiol. 1999 Oct;9(5):537-43. doi: 10.1016/S0959-4388(99)00020-3. Curr Opin Neurobiol. 1999. PMID: 10508746 Review.
-
Epigenetic Regulation of Notch Signaling During Drosophila Development.Adv Exp Med Biol. 2020;1218:59-75. doi: 10.1007/978-3-030-34436-8_4. Adv Exp Med Biol. 2020. PMID: 32060871 Review.
Cited by
-
Genomic insights into mite phylogeny, fitness, development, and reproduction.BMC Genomics. 2019 Dec 9;20(1):954. doi: 10.1186/s12864-019-6281-1. BMC Genomics. 2019. PMID: 31818245 Free PMC article.
-
HP1c regulates development and gut homeostasis by suppressing Notch signaling through Su(H).EMBO Rep. 2021 Apr 7;22(4):e51298. doi: 10.15252/embr.202051298. Epub 2021 Feb 17. EMBO Rep. 2021. PMID: 33594776 Free PMC article.
References
-
- Dexter J. S. The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. Am. Nat. 48, 712–758 (1914).
-
- Morgan T. H. B. & C. B. Sex-Linked Inheritance in Drosophila. PartII, 63–64 (Press of Gibson Brothers 1916).
-
- Wharton K. A., Johansen K. M., Xu T. & Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell 43, 567–581 (1985). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

