Mitochondrial genome evolution and tRNA truncation in Acariformes mites: new evidence from eriophyoid mites
- PMID: 26732998
- PMCID: PMC4702108
- DOI: 10.1038/srep18920
Mitochondrial genome evolution and tRNA truncation in Acariformes mites: new evidence from eriophyoid mites
Abstract
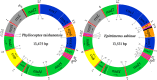
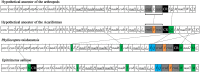
The subclass Acari (mites and ticks) comprises two super-orders: Acariformes and Parasitiformes. Most species of the Parasitiformes known retained the ancestral pattern of mitochondrial (mt) gene arrangement of arthropods, and their mt tRNAs have the typical cloverleaf structure. All of the species of the Acariformes known, however, have rearranged mt genomes and truncated mt tRNAs. We sequenced the mt genomes of two species of Eriophyoidea: Phyllocoptes taishanensis and Epitrimerus sabinae. The mt genomes of P. taishanensis and E. sabinae are 13,475 bp and 13,531 bp, respectively, are circular and contain the 37 genes typical of animals; most mt tRNAs are highly truncated in both mites. On the other hand, these two eriophyoid mites have the least rearranged mt genomes seen in the Acariformes. Comparison between eriophyoid mites and other Aacariformes mites showed that: 1) the most recent common ancestor of Acariformes mites retained the ancestral pattern of mt gene arrangement of arthropods with slight modifications; 2) truncation of tRNAs for cysteine, phenylalanine and histidine occurred once in the most recent common ancestor of Acariformes mites whereas truncation of other tRNAs occurred multiple times; and 3) the placement of eriophyoid mites in the order Trombidiformes needs to be reviewed.
Figures

Similar articles
-
The phylogenetic position of eriophyoid mites (superfamily Eriophyoidea) in Acariformes inferred from the sequences of mitochondrial genomes and nuclear small subunit (18S) rRNA gene.Mol Phylogenet Evol. 2017 Apr;109:271-282. doi: 10.1016/j.ympev.2017.01.009. Epub 2017 Jan 22. Mol Phylogenet Evol. 2017. PMID: 28119107
-
Complete mitochondrial genomes of the human follicle mites Demodex brevis and D. folliculorum: novel gene arrangement, truncated tRNA genes, and ancient divergence between species.BMC Genomics. 2014 Dec 16;15(1):1124. doi: 10.1186/1471-2164-15-1124. BMC Genomics. 2014. PMID: 25515815 Free PMC article.
-
Molecular mechanisms for the variation of mitochondrial gene content and gene arrangement among chigger mites of the genus Leptotrombidium (Acari: Acariformes).J Mol Evol. 2006 Aug;63(2):251-61. doi: 10.1007/s00239-005-0196-y. Epub 2006 Jul 7. J Mol Evol. 2006. PMID: 16830100
-
Where Eriophyoidea (Acariformes) Belong in the Tree of Life.Insects. 2023 Jun 6;14(6):527. doi: 10.3390/insects14060527. Insects. 2023. PMID: 37367343 Free PMC article. Review.
-
DNA-based methods for eriophyoid mite studies: review, critical aspects, prospects and challenges.Exp Appl Acarol. 2010 Jul;51(1-3):257-71. doi: 10.1007/s10493-009-9301-z. Epub 2009 Oct 14. Exp Appl Acarol. 2010. PMID: 19826904 Review.
Cited by
-
Suboral fork: a newly discerned gnathosomal structure from the proboscis of eriophyoid mites (Acari, Eriophyoidea).Exp Appl Acarol. 2016 Oct;70(2):137-53. doi: 10.1007/s10493-016-0077-7. Epub 2016 Aug 8. Exp Appl Acarol. 2016. PMID: 27502114
-
Unravelling the phylogeny, cryptic diversity and morphological evolution of Diptilomiopus mites (Acari: Eriophyoidea).Exp Appl Acarol. 2019 Dec;79(3-4):323-344. doi: 10.1007/s10493-019-00443-8. Epub 2019 Nov 30. Exp Appl Acarol. 2019. PMID: 31786687
-
Arm-less mitochondrial tRNAs conserved for over 30 millions of years in spiders.BMC Genomics. 2019 Aug 23;20(1):665. doi: 10.1186/s12864-019-6026-1. BMC Genomics. 2019. PMID: 31438844 Free PMC article.
-
Symbiotic bacteria of the gall-inducing mite Fragariocoptes setiger (Eriophyoidea) and phylogenomic resolution of the eriophyoid position among Acari.Sci Rep. 2022 Mar 9;12(1):3811. doi: 10.1038/s41598-022-07535-3. Sci Rep. 2022. PMID: 35264574 Free PMC article.
-
A new, sensitive and efficient method for taxonomic placement in the Eriophyoidea and virus detection in individual eriophyoids.Exp Appl Acarol. 2019 Jun;78(2):247-261. doi: 10.1007/s10493-019-00382-4. Epub 2019 May 25. Exp Appl Acarol. 2019. PMID: 31129764
References
-
- Zhang Z.-Q. in Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness (Magnolia Press, 2011). - PubMed
-
- Skoracka A., Smith L., Oldfield G., Cristofaro M. & Amrine J. W. Host-plant specificity and specialization in eriophyoid mites and their importance for the use of eriophyoid mites as biocontrol agents of weeds. Exp. Appl. Acarol. 51, 93–113 (2010). - PubMed
-
- Duso C., Castagnoli M., Simoni S. & Angeli G. The impact of eriophyoids on crops: recent issues on Aculus schlechtendali, Calepitrimerus vitis and Aculops lycopersici. Exp. Appl. Acarol. 51, 151–168 (2010). - PubMed
-
- Castagnoli M., Lewandowski M., Labanowski G. S., Simoni S. & Soika G. M. An insight into some relevant aspects concerning eriophyoid mites inhabiting forests, ornamental trees and shrubs. Exp. Appl. Acarol. 51, 169–189 (2010). - PubMed
-
- Miller A. D. et al. Phylogenetic analyses reveal extensive cryptic speciation and host specialization in an economically important mite taxon. Mol. Phylogenet. Evol. 66, 928–940 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

