Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma
- PMID: 26691987
- PMCID: PMC4731318
- DOI: 10.1038/ng.3473
Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma
Erratum in
-
Corrigendum: Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma.Nat Genet. 2016 May 27;48(6):700. doi: 10.1038/ng0616-700b. Nat Genet. 2016. PMID: 27230687 No abstract available.
Abstract
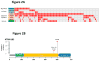
Follicular lymphoma is an incurable B cell malignancy characterized by the t(14;18) translocation and mutations affecting the epigenome. Although frequent gene mutations in key signaling pathways, including JAK-STAT, NOTCH and NF-κB, have also been defined, the spectrum of these mutations typically overlaps with that in the closely related diffuse large B cell lymphoma (DLBCL). Using a combination of discovery exome and extended targeted sequencing, we identified recurrent somatic mutations in RRAGC uniquely enriched in patients with follicular lymphoma (17%). More than half of the mutations preferentially co-occurred with mutations in ATP6V1B2 and ATP6AP1, which encode components of the vacuolar H(+)-ATP ATPase (V-ATPase) known to be necessary for amino acid-induced activation of mTORC1. The RagC variants increased raptor binding while rendering mTORC1 signaling resistant to amino acid deprivation. The activating nature of the RRAGC mutations, their existence in the dominant clone and their stability during disease progression support their potential as an excellent candidate for therapeutic targeting.
Conflict of interest statement
We declare no competing financial interests.
Figures




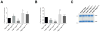
Similar articles
-
De novo RRAGC mutation activates mTORC1 signaling in syndromic fetal dilated cardiomyopathy.Hum Genet. 2016 Aug;135(8):909-917. doi: 10.1007/s00439-016-1685-3. Epub 2016 May 27. Hum Genet. 2016. PMID: 27234373 Free PMC article.
-
Recurrent Mutations in the MTOR Regulator RRAGC in Follicular Lymphoma.Clin Cancer Res. 2016 Nov 1;22(21):5383-5393. doi: 10.1158/1078-0432.CCR-16-0609. Epub 2016 Jun 7. Clin Cancer Res. 2016. PMID: 27267853 Free PMC article.
-
Raptor mediates the selective inhibitory effect of cardamonin on RRAGC-mutant B cell lymphoma.BMC Complement Med Ther. 2023 Sep 26;23(1):336. doi: 10.1186/s12906-023-04166-7. BMC Complement Med Ther. 2023. PMID: 37749558 Free PMC article.
-
Rag GTPase in amino acid signaling.Amino Acids. 2016 Apr;48(4):915-928. doi: 10.1007/s00726-016-2171-x. Epub 2016 Jan 18. Amino Acids. 2016. PMID: 26781224 Review.
-
Key mediators of intracellular amino acids signaling to mTORC1 activation.Amino Acids. 2015 May;47(5):857-67. doi: 10.1007/s00726-015-1937-x. Epub 2015 Feb 21. Amino Acids. 2015. PMID: 25701492 Review.
Cited by
-
The metabolic plasticity of B cells.Front Mol Biosci. 2022 Sep 23;9:991188. doi: 10.3389/fmolb.2022.991188. eCollection 2022. Front Mol Biosci. 2022. PMID: 36213123 Free PMC article. Review.
-
De novo RRAGC mutation activates mTORC1 signaling in syndromic fetal dilated cardiomyopathy.Hum Genet. 2016 Aug;135(8):909-917. doi: 10.1007/s00439-016-1685-3. Epub 2016 May 27. Hum Genet. 2016. PMID: 27234373 Free PMC article.
-
Emerging role of mTOR in the response to cancer therapeutics.Trends Cancer. 2016 May;2(5):241-251. doi: 10.1016/j.trecan.2016.03.008. Trends Cancer. 2016. PMID: 27668290 Free PMC article.
-
Metabolomics: A New Era in the Diagnosis or Prognosis of B-Cell Non-Hodgkin's Lymphoma.Diagnostics (Basel). 2023 Feb 23;13(5):861. doi: 10.3390/diagnostics13050861. Diagnostics (Basel). 2023. PMID: 36900005 Free PMC article. Review.
-
Integration of Mutational Signature Analysis with 3D Chromatin Data Unveils Differential AID-Related Mutagenesis in Indolent Lymphomas.Int J Mol Sci. 2021 Dec 1;22(23):13015. doi: 10.3390/ijms222313015. Int J Mol Sci. 2021. PMID: 34884820 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

