3D Chromosome Regulatory Landscape of Human Pluripotent Cells
- PMID: 26686465
- PMCID: PMC4848748
- DOI: 10.1016/j.stem.2015.11.007
3D Chromosome Regulatory Landscape of Human Pluripotent Cells
Abstract
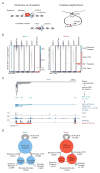
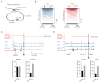
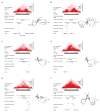
In this study, we describe the 3D chromosome regulatory landscape of human naive and primed embryonic stem cells. To devise this map, we identified transcriptional enhancers and insulators in these cells and placed them within the context of cohesin-associated CTCF-CTCF loops using cohesin ChIA-PET data. The CTCF-CTCF loops we identified form a chromosomal framework of insulated neighborhoods, which in turn form topologically associating domains (TADs) that are largely preserved during the transition between the naive and primed states. Regulatory changes in enhancer-promoter interactions occur within insulated neighborhoods during cell state transition. The CTCF anchor regions we identified are conserved across species, influence gene expression, and are a frequent site of mutations in cancer cells, underscoring their functional importance in cellular regulation. These 3D regulatory maps of human pluripotent cells therefore provide a foundation for future interrogation of the relationships between chromosome structure and gene control in development and disease.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Naive versus Primed: It's Now Three-Dimensional.Cell Stem Cell. 2016 Feb 4;18(2):164-5. doi: 10.1016/j.stem.2016.01.014. Cell Stem Cell. 2016. PMID: 26849300
Similar articles
-
The long-range interaction landscape of gene promoters.Nature. 2012 Sep 6;489(7414):109-13. doi: 10.1038/nature11279. Nature. 2012. PMID: 22955621 Free PMC article.
-
Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo.Nat Cell Biol. 2017 Aug;19(8):952-961. doi: 10.1038/ncb3573. Epub 2017 Jul 24. Nat Cell Biol. 2017. PMID: 28737770 Free PMC article.
-
Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes.Cell. 2014 Oct 9;159(2):374-387. doi: 10.1016/j.cell.2014.09.030. Cell. 2014. PMID: 25303531 Free PMC article.
-
Insulators and domains of gene expression.Curr Opin Genet Dev. 2016 Apr;37:17-26. doi: 10.1016/j.gde.2015.11.009. Epub 2016 Jan 20. Curr Opin Genet Dev. 2016. PMID: 26802288 Review.
-
Function and regulation of chromatin insulators in dynamic genome organization.Curr Opin Cell Biol. 2019 Jun;58:61-68. doi: 10.1016/j.ceb.2019.02.001. Epub 2019 Mar 12. Curr Opin Cell Biol. 2019. PMID: 30875678 Free PMC article. Review.
Cited by
-
Widespread reorganisation of pluripotent factor binding and gene regulatory interactions between human pluripotent states.Nat Commun. 2021 Apr 7;12(1):2098. doi: 10.1038/s41467-021-22201-4. Nat Commun. 2021. PMID: 33828098 Free PMC article.
-
Transcription factors: building hubs in the 3D space.Cell Cycle. 2020 Oct;19(19):2395-2410. doi: 10.1080/15384101.2020.1805238. Epub 2020 Aug 12. Cell Cycle. 2020. PMID: 32783593 Free PMC article. Review.
-
Deletion of CTCF sites in the SHH locus alters enhancer-promoter interactions and leads to acheiropodia.Nat Commun. 2021 Apr 16;12(1):2282. doi: 10.1038/s41467-021-22470-z. Nat Commun. 2021. PMID: 33863876 Free PMC article.
-
Chromosome conformation capture approaches to investigate 3D genome architecture in Ankylosing Spondylitis.Front Genet. 2023 Jan 25;14:1129207. doi: 10.3389/fgene.2023.1129207. eCollection 2023. Front Genet. 2023. PMID: 36760998 Free PMC article.
-
Control of directionality of chromatin folding for the inter- and intra-domain contacts at the Tfap2c-Bmp7 locus.Epigenetics Chromatin. 2018 Sep 14;11(1):51. doi: 10.1186/s13072-018-0221-1. Epigenetics Chromatin. 2018. PMID: 30213272 Free PMC article.
References
-
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA-sequences. Cell. 1981;27:299–308. - PubMed
-
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
-
- Benoist C, Chambon P. In vivo sequence requirements of the SV40 early promoter region. Nature. 1981;290:304–310. - PubMed
-
- Bickmore WA. The Spatial Organization of the Human Genome. Annual Review of Genomics and Human Genetics. 2013;14:67–84. - PubMed
-
- Calhoun ES, Jones JB, Ashfaq R, Adsay V, Baker SJ, Valentine V, Hempen PM, Hilgers W, Yeo CJ, Hruban RH, et al. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. The American journal of pathology. 2003;163:1255–1260. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

