TET-catalyzed 5-hydroxymethylcytosine regulates gene expression in differentiating colonocytes and colon cancer
- PMID: 26631571
- PMCID: PMC4668370
- DOI: 10.1038/srep17568
TET-catalyzed 5-hydroxymethylcytosine regulates gene expression in differentiating colonocytes and colon cancer
Erratum in
-
Corrigendum: TET-catalyzed 5-hydroxymethylcytosine regulates gene expression in differentiating colonocytes and colon cancer.Sci Rep. 2016 Apr 28;6:24963. doi: 10.1038/srep24963. Sci Rep. 2016. PMID: 27121680 Free PMC article. No abstract available.
Abstract
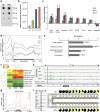
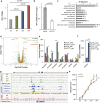
The formation of differentiated cell types from pluripotent progenitors involves epigenetic regulation of gene expression. DNA hydroxymethylation results from the enzymatic oxidation of 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) by the ten-eleven translocation (TET) 5-mC dioxygenase enzymes. Previous work has mapped changes in 5-mC during differentiation of intestinal stem cells. However, whether or not 5-hmC regulates colonocyte differentiation is unknown. Here we show that 5-hmC regulates gene expression during colonocyte differentiation and controls gene expression in human colon cancers. Genome-wide profiling of 5-hmC during in vitro colonic differentiation demonstrated that 5-hmC is gained at highly expressed and induced genes and is associated with intestinal transcription factor binding sites, including those for HNF4A and CDX2. TET1 induction occurred during differentiation, and TET1 knockdown altered gene expression and inhibited barrier formation of colonocytes. We find that the 5-hmC distribution in primary human colonocytes parallels the distribution found in differentiated cells in vitro, and that gene-specific 5-hmC changes in human colon cancers are directly correlated with changes in gene expression. Our results support a model in which 5-hmC regulates differentiation of adult human intestine and 5-hmC alterations contribute to the disrupted gene expression in colon cancer.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine.J Biol Chem. 2013 May 10;288(19):13669-74. doi: 10.1074/jbc.C113.464800. Epub 2013 Apr 2. J Biol Chem. 2013. PMID: 23548903 Free PMC article.
-
TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma.Cell Rep. 2014 Jun 12;7(5):1343-1352. doi: 10.1016/j.celrep.2014.04.040. Epub 2014 May 15. Cell Rep. 2014. PMID: 24835990 Free PMC article.
-
Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1.PLoS One. 2013 May 9;8(5):e62828. doi: 10.1371/journal.pone.0062828. Print 2013. PLoS One. 2013. PMID: 23671639 Free PMC article.
-
Ten-eleven translocation proteins (TETs): tumor suppressors or tumor enhancers?Front Biosci (Landmark Ed). 2021 Oct 30;26(10):895-915. doi: 10.52586/4996. Front Biosci (Landmark Ed). 2021. PMID: 34719214 Review.
-
Ten-eleven translocase: key regulator of the methylation landscape in cancer.J Cancer Res Clin Oncol. 2021 Jul;147(7):1869-1879. doi: 10.1007/s00432-021-03641-3. Epub 2021 Apr 28. J Cancer Res Clin Oncol. 2021. PMID: 33913031 Review.
Cited by
-
Bronchial Epithelial Tet2 Maintains Epithelial Integrity during Acute Pseudomonas aeruginosa Pneumonia.Infect Immun. 2020 Dec 15;89(1):e00603-20. doi: 10.1128/IAI.00603-20. Print 2020 Dec 15. Infect Immun. 2020. PMID: 33046509 Free PMC article.
-
DNA Hydroxymethylation in Smoking-Associated Cancers.Int J Mol Sci. 2022 Feb 28;23(5):2657. doi: 10.3390/ijms23052657. Int J Mol Sci. 2022. PMID: 35269796 Free PMC article. Review.
-
TET1 Depletion Induces Aberrant CpG Methylation in Colorectal Cancer Cells.PLoS One. 2016 Dec 15;11(12):e0168281. doi: 10.1371/journal.pone.0168281. eCollection 2016. PLoS One. 2016. PMID: 27977763 Free PMC article.
-
Impact of polymorphisms within genes involved in regulating DNA methylation in patients with metastatic colorectal cancer enrolled in three independent, randomised, open-label clinical trials: a meta-analysis from TRIBE, MAVERICC and FIRE-3.Eur J Cancer. 2019 Apr;111:138-147. doi: 10.1016/j.ejca.2019.01.105. Epub 2019 Mar 7. Eur J Cancer. 2019. PMID: 30852420 Free PMC article.
-
Hypercholesterolemia Increases Colorectal Cancer Incidence by Reducing Production of NKT and γδ T Cells from Hematopoietic Stem Cells.Cancer Res. 2017 May 1;77(9):2351-2362. doi: 10.1158/0008-5472.CAN-16-1916. Epub 2017 Mar 1. Cancer Res. 2017. PMID: 28249902 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

