The Transcription and Translation Landscapes during Human Cytomegalovirus Infection Reveal Novel Host-Pathogen Interactions
- PMID: 26599541
- PMCID: PMC4658056
- DOI: 10.1371/journal.ppat.1005288
The Transcription and Translation Landscapes during Human Cytomegalovirus Infection Reveal Novel Host-Pathogen Interactions
Abstract
Viruses are by definition fully dependent on the cellular translation machinery, and develop diverse mechanisms to co-opt this machinery for their own benefit. Unlike many viruses, human cytomegalovirus (HCMV) does suppress the host translation machinery, and the extent to which translation machinery contributes to the overall pattern of viral replication and pathogenesis remains elusive. Here, we combine RNA sequencing and ribosomal profiling analyses to systematically address this question. By simultaneously examining the changes in transcription and translation along HCMV infection, we uncover extensive transcriptional control that dominates the response to infection, but also diverse and dynamic translational regulation for subsets of host genes. We were also able to show that, at late time points in infection, translation of viral mRNAs is higher than that of cellular mRNAs. Lastly, integration of our translation measurements with recent measurements of protein abundance enabled comprehensive identification of dozens of host proteins that are targeted for degradation during HCMV infection. Since targeted degradation indicates a strong biological importance, this approach should be applicable for discovering central host functions during viral infection. Our work provides a framework for studying the contribution of transcription, translation and degradation during infection with any virus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
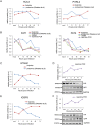

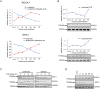
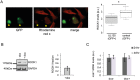
Similar articles
-
Differential role for host translation factors in host and viral protein synthesis during human cytomegalovirus infection.J Virol. 2014 Feb;88(3):1473-83. doi: 10.1128/JVI.02321-13. Epub 2013 Nov 6. J Virol. 2014. PMID: 24198422 Free PMC article.
-
Human Cytomegalovirus Strategies to Maintain and Promote mRNA Translation.Viruses. 2016 Apr 13;8(4):97. doi: 10.3390/v8040097. Viruses. 2016. PMID: 27089357 Free PMC article. Review.
-
Human cytomegalovirus TRS1 protein associates with the 7-methylguanosine mRNA cap and facilitates translation.Proteomics. 2015 Jun;15(12):1983-94. doi: 10.1002/pmic.201400616. Epub 2015 May 12. Proteomics. 2015. PMID: 25894605 Free PMC article.
-
The 5' Untranslated Region of the Major Immediate Early mRNA Is Necessary for Efficient Human Cytomegalovirus Replication.J Virol. 2018 Mar 14;92(7):e02128-17. doi: 10.1128/JVI.02128-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29343581 Free PMC article.
-
MicroRNAs expressed by human cytomegalovirus.Virol J. 2020 Mar 12;17(1):34. doi: 10.1186/s12985-020-1296-4. Virol J. 2020. PMID: 32164742 Free PMC article. Review.
Cited by
-
Identification of UL69 Gene and Protein in Cytomegalovirus-Transformed Human Mammary Epithelial Cells.Front Oncol. 2021 Apr 16;11:627866. doi: 10.3389/fonc.2021.627866. eCollection 2021. Front Oncol. 2021. PMID: 33937031 Free PMC article.
-
Control of animal virus replication by RNA adenosine methylation.Adv Virus Res. 2022;112:87-114. doi: 10.1016/bs.aivir.2022.01.002. Epub 2022 Mar 7. Adv Virus Res. 2022. PMID: 35840182 Free PMC article. Review.
-
Human Cytomegalovirus in breast milk is associated with milk composition, the infant gut microbiome, and infant growth.bioRxiv [Preprint]. 2023 Jul 19:2023.07.19.549370. doi: 10.1101/2023.07.19.549370. bioRxiv. 2023. Update in: Nat Commun. 2024 Jul 23;15(1):6216. doi: 10.1038/s41467-024-50282-4 PMID: 37503212 Free PMC article. Updated. Preprint.
-
Translational Control in Virus-Infected Cells.Cold Spring Harb Perspect Biol. 2019 Mar 1;11(3):a033001. doi: 10.1101/cshperspect.a033001. Cold Spring Harb Perspect Biol. 2019. PMID: 29891561 Free PMC article. Review.
-
Epigenetic reprogramming of host and viral genes by Human Cytomegalovirus infection in Kasumi-3 myeloid progenitor cells at early times post-infection.J Virol. 2021 May 10;95(11):e00183-21. doi: 10.1128/JVI.00183-21. Epub 2021 Mar 17. J Virol. 2021. PMID: 33731453 Free PMC article.
References
-
- Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, et al. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol. 2003/01/21 ed. 2003;84: 17–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

