The multifaceted roles of the HORMA domain in cellular signaling
- PMID: 26598612
- PMCID: PMC4657174
- DOI: 10.1083/jcb.201509076
The multifaceted roles of the HORMA domain in cellular signaling
Abstract
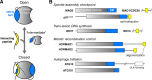
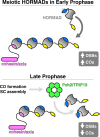
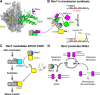
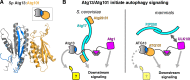
The HORMA domain is a multifunctional protein-protein interaction module found in diverse eukaryotic signaling pathways including the spindle assembly checkpoint, numerous DNA recombination/repair pathways, and the initiation of autophagy. In all of these pathways, HORMA domain proteins occupy key signaling junctures and function through the controlled assembly and disassembly of signaling complexes using a stereotypical "safety belt" peptide interaction mechanism. A recent explosion of structural and functional work has shed new light on these proteins, illustrating how strikingly similar structural mechanisms give rise to radically different functional outcomes in each family of HORMA domain proteins.
© 2015 Rosenberg and Corbett.
Figures

Similar articles
-
Conformational dynamics of the Hop1 HORMA domain reveal a common mechanism with the spindle checkpoint protein Mad2.Nucleic Acids Res. 2018 Jan 9;46(1):279-292. doi: 10.1093/nar/gkx1196. Nucleic Acids Res. 2018. PMID: 29186573 Free PMC article.
-
The HORMA domain: an evolutionarily conserved domain discovered in chromatin-associated proteins, has unanticipated diverse functions.Gene. 2014 Jul 25;545(2):194-7. doi: 10.1016/j.gene.2014.05.020. Epub 2014 May 9. Gene. 2014. PMID: 24814187 Review.
-
A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy.Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5486-91. doi: 10.1073/pnas.1220306110. Epub 2013 Mar 18. Proc Natl Acad Sci U S A. 2013. PMID: 23509291 Free PMC article.
-
Evolutionary Dynamics and Molecular Mechanisms of HORMA Domain Protein Signaling.Annu Rev Biochem. 2022 Jun 21;91:541-569. doi: 10.1146/annurev-biochem-090920-103246. Epub 2022 Jan 18. Annu Rev Biochem. 2022. PMID: 35041460 Review.
-
The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair.Trends Biochem Sci. 1998 Aug;23(8):284-6. doi: 10.1016/s0968-0004(98)01257-2. Trends Biochem Sci. 1998. PMID: 9757827 Review. No abstract available.
Cited by
-
Does the Pachytene Checkpoint, a Feature of Meiosis, Filter Out Mistakes in Double-Strand DNA Break Repair and as a side-Effect Strongly Promote Adaptive Speciation?Integr Org Biol. 2022 Apr 8;4(1):obac008. doi: 10.1093/iob/obac008. eCollection 2022. Integr Org Biol. 2022. PMID: 36827645 Free PMC article.
-
Novel mechanistic insights into the role of Mer2 as the keystone of meiotic DNA break formation.Elife. 2021 Dec 24;10:e72330. doi: 10.7554/eLife.72330. Elife. 2021. PMID: 34951404 Free PMC article.
-
Genetic interactions between the chromosome axis-associated protein Hop1 and homologous recombination determinants in Schizosaccharomyces pombe.Curr Genet. 2018 Oct;64(5):1089-1104. doi: 10.1007/s00294-018-0827-7. Epub 2018 Mar 17. Curr Genet. 2018. PMID: 29550859 Free PMC article.
-
The ectopic expression of meiCT genes promotes meiomitosis and may facilitate carcinogenesis.Cell Cycle. 2020 Apr;19(8):837-854. doi: 10.1080/15384101.2020.1743902. Epub 2020 Mar 30. Cell Cycle. 2020. PMID: 32223693 Free PMC article. Review.
-
The Arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis.EMBO J. 2020 Feb 3;39(3):e101625. doi: 10.15252/embj.2019101625. Epub 2019 Sep 26. EMBO J. 2020. PMID: 31556459 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

