SureChEMBL: a large-scale, chemically annotated patent document database
- PMID: 26582922
- PMCID: PMC4702887
- DOI: 10.1093/nar/gkv1253
SureChEMBL: a large-scale, chemically annotated patent document database
Abstract
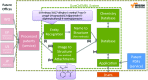
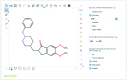
SureChEMBL is a publicly available large-scale resource containing compounds extracted from the full text, images and attachments of patent documents. The data are extracted from the patent literature according to an automated text and image-mining pipeline on a daily basis. SureChEMBL provides access to a previously unavailable, open and timely set of annotated compound-patent associations, complemented with sophisticated combined structure and keyword-based search capabilities against the compound repository and patent document corpus; given the wealth of knowledge hidden in patent documents, analysis of SureChEMBL data has immediate applications in drug discovery, medicinal chemistry and other commercial areas of chemical science. Currently, the database contains 17 million compounds extracted from 14 million patent documents. Access is available through a dedicated web-based interface and data downloads at: https://www.surechembl.org/.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Exploring SureChEMBL from a drug discovery perspective.Sci Data. 2024 May 16;11(1):507. doi: 10.1038/s41597-024-03371-4. Sci Data. 2024. PMID: 38755219 Free PMC article.
-
Managing expectations: assessment of chemistry databases generated by automated extraction of chemical structures from patents.J Cheminform. 2015 Oct 6;7(1):49. doi: 10.1186/s13321-015-0097-z. eCollection 2015 Dec. J Cheminform. 2015. PMID: 26457120 Free PMC article.
-
Chemical named entity recognition in patents by domain knowledge and unsupervised feature learning.Database (Oxford). 2016 Apr 17;2016:baw049. doi: 10.1093/database/baw049. Print 2016. Database (Oxford). 2016. PMID: 27087307 Free PMC article.
-
Expanding opportunities for mining bioactive chemistry from patents.Drug Discov Today Technol. 2015 Jul;14:3-9. doi: 10.1016/j.ddtec.2014.12.001. Epub 2015 Feb 11. Drug Discov Today Technol. 2015. PMID: 26194581 Free PMC article. Review.
-
Opening up connectivity between documents, structures and bioactivity.Beilstein J Org Chem. 2020 Apr 2;16:596-606. doi: 10.3762/bjoc.16.54. eCollection 2020. Beilstein J Org Chem. 2020. PMID: 32280387 Free PMC article. Review.
Cited by
-
Open PHACTS computational protocols for in silico target validation of cellular phenotypic screens: knowing the knowns.Medchemcomm. 2016 Jun 1;7(6):1237-1244. doi: 10.1039/c6md00065g. Epub 2016 May 11. Medchemcomm. 2016. PMID: 27774140 Free PMC article.
-
Using alternative SMILES representations to identify novel functional analogues in chemical similarity vector searches.Patterns (N Y). 2023 Oct 30;4(12):100865. doi: 10.1016/j.patter.2023.100865. eCollection 2023 Dec 8. Patterns (N Y). 2023. PMID: 38106612 Free PMC article.
-
Scaffold-Hopping from Synthetic Drugs by Holistic Molecular Representation.Sci Rep. 2018 Nov 7;8(1):16469. doi: 10.1038/s41598-018-34677-0. Sci Rep. 2018. PMID: 30405170 Free PMC article.
-
Successive Statistical and Structure-Based Modeling to Identify Chemically Novel Kinase Inhibitors.J Chem Inf Model. 2020 Sep 28;60(9):4283-4295. doi: 10.1021/acs.jcim.9b01204. Epub 2020 May 12. J Chem Inf Model. 2020. PMID: 32343143 Free PMC article.
-
Using ChEMBL web services for building applications and data processing workflows relevant to drug discovery.Expert Opin Drug Discov. 2017 Aug;12(8):757-767. doi: 10.1080/17460441.2017.1339032. Epub 2017 Jun 12. Expert Opin Drug Discov. 2017. PMID: 28602100 Free PMC article. Review.
References
-
- Downs G.M., Barnard J.M. Chemical patent information systems. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011;1:727–741.
-
- Bregonje M. Patents: a unique source for scientific technical information in chemistry related industry. World Patent Inf. 2005;27:309–315.
-
- Schneider N., Lowe D.M., Sayle R.A., Landrum G.A. Development of a novel fingerprint for chemical reactions and its application to large-scale reaction classification and similarity. J. Chem. Inf. Model. 2015;55:39–53. - PubMed
-
- Kettle J.G., Ward R.A., Griffen E. Data-mining patent literature for novel chemical reagents for use in medicinal chemistry design. Medchemcomm. 2010;1:331–338.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

