C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir
- PMID: 26581995
- PMCID: PMC4810716
- DOI: 10.1128/JVI.01829-15
C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir
Abstract
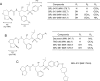
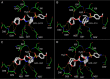
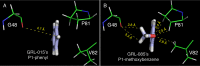
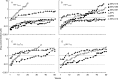
We identified three nonpeptidic HIV-1 protease inhibitors (PIs), GRL-015, -085, and -097, containing tetrahydropyrano-tetrahydrofuran (Tp-THF) with a C-5 hydroxyl. The three compounds were potent against a wild-type laboratory HIV-1 strain (HIV-1(WT)), with 50% effective concentrations (EC50s) of 3.0 to 49 nM, and exhibited minimal cytotoxicity, with 50% cytotoxic concentrations (CC50) for GRL-015, -085, and -097 of 80, >100, and >100 μM, respectively. All the three compounds potently inhibited the replication of highly PI-resistant HIV-1 variants selected with each of the currently available PIs and recombinant clinical HIV-1 isolates obtained from patients harboring multidrug-resistant HIV-1 variants (HIVMDR). Importantly, darunavir (DRV) was >1,000 times less active against a highly DRV-resistant HIV-1 variant (HIV-1DRV(R) P51); the three compounds remained active against HIV-1DRV(R) P51 with only a 6.8- to 68-fold reduction. Moreover, the emergence of HIV-1 variants resistant to the three compounds was considerably delayed compared to the case of DRV. In particular, HIV-1 variants resistant to GRL-085 and -097 did not emerge even when two different highly DRV-resistant HIV-1 variants were used as a starting population. In the structural analyses, Tp-THF of GRL-015, -085, and -097 showed strong hydrogen bond interactions with the backbone atoms of active-site amino acid residues (Asp29 and Asp30) of HIV-1 protease. A strong hydrogen bonding formation between the hydroxyl moiety of Tp-THF and a carbonyl oxygen atom of Gly48 was newly identified. The present findings indicate that the three compounds warrant further study as possible therapeutic agents for treating individuals harboring wild-type HIV and/or HIVMDR.
Importance: Darunavir (DRV) inhibits the replication of most existing multidrug-resistant HIV-1 strains and has a high genetic barrier. However, the emergence of highly DRV-resistant HIV-1 strains (HIVDRV(R) ) has recently been observed in vivo and in vitro. Here, we identified three novel HIV-1 protease inhibitors (PIs) containing a tetrahydropyrano-tetrahydrofuran (Tp-THF) moiety with a C-5 hydroxyl (GRL-015, -085, and -097) which potently suppress the replication of HIVDRV(R) . Moreover, the emergence of HIV-1 strains resistant to the three compounds was considerably delayed compared to the case of DRV. The C-5 hydroxyl formed a strong hydrogen bonding interaction with the carbonyl oxygen atom of Gly48 of protease as examined in the structural analyses. Interestingly, a compound with Tp-THF lacking the hydroxyl moiety substantially decreased activity against HIVDRV(R) . The three novel compounds should be further developed as potential drugs for treating individuals harboring wild-type and multi-PI-resistant HIV variants as well as HIVDRV(R) .
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
GRL-079, a Novel HIV-1 Protease Inhibitor, Is Extremely Potent against Multidrug-Resistant HIV-1 Variants and Has a High Genetic Barrier against the Emergence of Resistant Variants.Antimicrob Agents Chemother. 2018 Apr 26;62(5):e02060-17. doi: 10.1128/AAC.02060-17. Print 2018 May. Antimicrob Agents Chemother. 2018. PMID: 29463535 Free PMC article.
-
Novel Protease Inhibitors Containing C-5-Modified bis-Tetrahydrofuranylurethane and Aminobenzothiazole as P2 and P2' Ligands That Exert Potent Antiviral Activity against Highly Multidrug-Resistant HIV-1 with a High Genetic Barrier against the Emergence of Drug Resistance.Antimicrob Agents Chemother. 2019 Jul 25;63(8):e00372-19. doi: 10.1128/AAC.00372-19. Print 2019 Aug. Antimicrob Agents Chemother. 2019. PMID: 31085520 Free PMC article.
-
GRL-0519, a novel oxatricyclic ligand-containing nonpeptidic HIV-1 protease inhibitor (PI), potently suppresses replication of a wide spectrum of multi-PI-resistant HIV-1 variants in vitro.Antimicrob Agents Chemother. 2013 May;57(5):2036-46. doi: 10.1128/AAC.02189-12. Epub 2013 Feb 12. Antimicrob Agents Chemother. 2013. PMID: 23403426 Free PMC article.
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
Cited by
-
GRL-079, a Novel HIV-1 Protease Inhibitor, Is Extremely Potent against Multidrug-Resistant HIV-1 Variants and Has a High Genetic Barrier against the Emergence of Resistant Variants.Antimicrob Agents Chemother. 2018 Apr 26;62(5):e02060-17. doi: 10.1128/AAC.02060-17. Print 2018 May. Antimicrob Agents Chemother. 2018. PMID: 29463535 Free PMC article.
-
Novel Protease Inhibitors Containing C-5-Modified bis-Tetrahydrofuranylurethane and Aminobenzothiazole as P2 and P2' Ligands That Exert Potent Antiviral Activity against Highly Multidrug-Resistant HIV-1 with a High Genetic Barrier against the Emergence of Drug Resistance.Antimicrob Agents Chemother. 2019 Jul 25;63(8):e00372-19. doi: 10.1128/AAC.00372-19. Print 2019 Aug. Antimicrob Agents Chemother. 2019. PMID: 31085520 Free PMC article.
-
A Photochemical Route to Optically Active Hexahydro-4H-furopyranol, a High-Affinity P2 Ligand for HIV-1 Protease Inhibitors.J Org Chem. 2019 Aug 2;84(15):9801-9805. doi: 10.1021/acs.joc.9b01361. Epub 2019 Jul 16. J Org Chem. 2019. PMID: 31310117 Free PMC article.
-
Beyond darunavir: recent development of next generation HIV-1 protease inhibitors to combat drug resistance.Chem Commun (Camb). 2022 Oct 20;58(84):11762-11782. doi: 10.1039/d2cc04541a. Chem Commun (Camb). 2022. PMID: 36200462 Free PMC article. Review.
-
Identification of Darunavir Derivatives for Inhibition of SARS-CoV-2 3CLpro.Int J Mol Sci. 2022 Dec 16;23(24):16011. doi: 10.3390/ijms232416011. Int J Mol Sci. 2022. PMID: 36555652 Free PMC article.
References
-
- Edmonds A, Yotebieng M, Lusiama J, Matumona Y, Kitetele F, Napravnik S, Cole SR, Van Rie A, Behets F. 2011. The effect of highly active antiretroviral therapy on the survival of HIV-infected children in a resource-deprived setting: a cohort study. PLoS Med 8:e1001044. doi:10.1371/journal.pmed.1001044. - DOI - PMC - PubMed
-
- Tejerina F, Bernaldo de Quiros JC. 2011. Protease inhibitors as preferred initial regimen for antiretroviral-naive HIV patients. AIDS Rev 13:227–233. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

