MIR517C inhibits autophagy and the epithelial-to-mesenchymal (-like) transition phenotype in human glioblastoma through KPNA2-dependent disruption of TP53 nuclear translocation
- PMID: 26553592
- PMCID: PMC4835194
- DOI: 10.1080/15548627.2015.1108507
MIR517C inhibits autophagy and the epithelial-to-mesenchymal (-like) transition phenotype in human glioblastoma through KPNA2-dependent disruption of TP53 nuclear translocation
Erratum in
-
Corrigendum.Autophagy. 2016;12(2):445-8. doi: 10.1080/15548627.2016.1137171. Autophagy. 2016. PMID: 26902592 Free PMC article. No abstract available.
-
Correction.Autophagy. 2019 Jun;15(6):1125-1127. doi: 10.1080/15548627.2019.1599190. Epub 2019 Mar 28. Autophagy. 2019. PMID: 30920334 Free PMC article. No abstract available.
Abstract
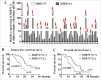
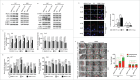
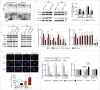
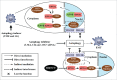
The epithelial-to-mesenchymal (-like) transition (EMT), a crucial embryonic development program, has been linked to the regulation of glioblastoma (GBM) progression and invasion. Here, we investigated the role of MIR517C/miR-517c, which belongs to the C19MC microRNA cluster identified in our preliminary studies, in the pathogenesis of GBM. We found that MIR517C was associated with improved prognosis in patients with GBM. Furthermore, following treatment with the autophagy inducer temozolomide (TMZ) and low glucose (LG), MIR517C degraded KPNA2 (karyopherin alpha 2 [RAG cohort 1, importin alpha 1]) and subsequently disturbed the nuclear translocation of TP53 in the GBM cell line U87 in vitro. Interestingly, this microRNA could inhibit autophagy and reduce cell migration and infiltration in U87 cells harboring wild-type (WT) TP53, but not in U251 cells harboring mutant (MU) TP53. Moreover, the expression of epithelial markers (i.e., CDH13/T-cadherin and CLDN1 [claudin 1]) increased, while the expression of mesenchymal markers (i.e., CDH2/N-cadherin, SNAI1/Snail, and VIM [vimentin]) decreased, indicating that the EMT status was blocked by MIR517C in U87 cells. Compared with MIR517C overexpression, MIR517C knockdown promoted infiltration of U87 cells to the surrounding structures in nude mice in vivo. The above phenotypic changes were also observed in TP53(+/+) and TP53(-/-) HCT116 colon cancer cells. In summary, our study provided support for a link between autophagy and EMT status in WT TP53 GBM cells and provided evidence for the signaling pathway (MIR517C-KPNA2-cytoplasmic TP53) involved in attenuating autophagy and eliminating the increased migration and invasion during the EMT.
Keywords: TP53; autophagy; epithelial-to-mesenchymal (-like) transition; glioblastoma; microRNA.
Figures







Similar articles
-
MicroRNA-128-3p Enhances the Chemosensitivity of Temozolomide in Glioblastoma by Targeting c-Met and EMT.Sci Rep. 2020 Jun 11;10(1):9471. doi: 10.1038/s41598-020-65331-3. Sci Rep. 2020. PMID: 32528036 Free PMC article.
-
Targeting the Notch1 oncogene by miR-139-5p inhibits glioma metastasis and epithelial-mesenchymal transition (EMT).BMC Neurol. 2018 Aug 31;18(1):133. doi: 10.1186/s12883-018-1139-8. BMC Neurol. 2018. PMID: 30170559 Free PMC article.
-
Reciprocal regulation of the cholinic phenotype and epithelial-mesenchymal transition in glioblastoma cells.Oncotarget. 2016 Nov 8;7(45):73414-73431. doi: 10.18632/oncotarget.12337. Oncotarget. 2016. PMID: 27705917 Free PMC article.
-
EMT Regulation by Autophagy: A New Perspective in Glioblastoma Biology.Cancers (Basel). 2019 Mar 6;11(3):312. doi: 10.3390/cancers11030312. Cancers (Basel). 2019. PMID: 30845654 Free PMC article. Review.
-
Multifaceted WNT Signaling at the Crossroads Between Epithelial-Mesenchymal Transition and Autophagy in Glioblastoma.Front Oncol. 2020 Nov 12;10:597743. doi: 10.3389/fonc.2020.597743. eCollection 2020. Front Oncol. 2020. PMID: 33312955 Free PMC article. Review.
Cited by
-
IRF2 Destabilizes Oncogenic KPNA2 to Modulate the Development of Osteosarcoma.J Oncol. 2022 Sep 26;2022:9973519. doi: 10.1155/2022/9973519. eCollection 2022. J Oncol. 2022. PMID: 36199790 Free PMC article.
-
HMGB1-Induced p62 Overexpression Promotes Snail-Mediated Epithelial-Mesenchymal Transition in Glioblastoma Cells via the Degradation of GSK-3β.Theranostics. 2019 Mar 16;9(7):1909-1922. doi: 10.7150/thno.30578. eCollection 2019. Theranostics. 2019. PMID: 31037147 Free PMC article.
-
MicroRNA-139 inhibits hepatocellular carcinoma cell growth through down-regulating karyopherin alpha 2.J Exp Clin Cancer Res. 2019 May 2;38(1):182. doi: 10.1186/s13046-019-1175-2. J Exp Clin Cancer Res. 2019. PMID: 31046781 Free PMC article.
-
Mitofusin1 Is a Major Mediator in Glucose-Induced Epithelial-to-Mesenchymal Transition in Lung Adenocarcinoma Cells.Onco Targets Ther. 2020 Apr 24;13:3511-3523. doi: 10.2147/OTT.S238714. eCollection 2020. Onco Targets Ther. 2020. PMID: 32425551 Free PMC article.
-
Decorin-mediated inhibition of the migration of U87MG glioma cells involves activation of autophagy and suppression of TGF-β signaling.FEBS Open Bio. 2016 May 31;6(7):707-19. doi: 10.1002/2211-5463.12076. eCollection 2016 Jul. FEBS Open Bio. 2016. PMID: 27398310 Free PMC article.
References
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al.. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352:987-96; PMID:15758009; http://dx.doi.org/10.1056/NEJMoa043330 - DOI - PubMed
-
- Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, Knuechel R, Kirchner T. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A 2001; 98:10356-61; PMID:11526241; http://dx.doi.org/10.1073/pnas.171610498 - DOI - PMC - PubMed
-
- Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer 2005; 5:744-9; PMID:16148886; http://dx.doi.org/10.1038/nrc1694 - DOI - PubMed
-
- Kahlert UD, Nikkhah G, Maciaczyk J. Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett 2013; 331:131-8; PMID:23268331; http://dx.doi.org/10.1016/j.canlet.2012.12.010 - DOI - PubMed
-
- Azab AK, Hu J, Quang P, Azab F, Pitsillides C, Awwad R, Thompson B, Maiso P, Sun JD, Hart CP, et al.. Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition-like features. Blood 2012; 119:5782-94; PMID:22394600; http://dx.doi.org/10.1182/blood-2011-09-380410 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
