Tools and data services registry: a community effort to document bioinformatics resources
- PMID: 26538599
- PMCID: PMC4702812
- DOI: 10.1093/nar/gkv1116
Tools and data services registry: a community effort to document bioinformatics resources
Abstract
Life sciences are yielding huge data sets that underpin scientific discoveries fundamental to improvement in human health, agriculture and the environment. In support of these discoveries, a plethora of databases and tools are deployed, in technically complex and diverse implementations, across a spectrum of scientific disciplines. The corpus of documentation of these resources is fragmented across the Web, with much redundancy, and has lacked a common standard of information. The outcome is that scientists must often struggle to find, understand, compare and use the best resources for the task at hand.Here we present a community-driven curation effort, supported by ELIXIR-the European infrastructure for biological information-that aspires to a comprehensive and consistent registry of information about bioinformatics resources. The sustainable upkeep of this Tools and Data Services Registry is assured by a curation effort driven by and tailored to local needs, and shared amongst a network of engaged partners.As of November 2015, the registry includes 1785 resources, with depositions from 126 individual registrations including 52 institutional providers and 74 individuals. With community support, the registry can become a standard for dissemination of information about bioinformatics resources: we welcome everyone to join us in this common endeavour. The registry is freely available at https://bio.tools.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
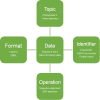
Similar articles
-
Community curation of bioinformatics software and data resources.Brief Bioinform. 2020 Sep 25;21(5):1697-1705. doi: 10.1093/bib/bbz075. Brief Bioinform. 2020. PMID: 31624831 Free PMC article.
-
Resource Disambiguator for the Web: Extracting Biomedical Resources and Their Citations from the Scientific Literature.PLoS One. 2016 Jan 5;11(1):e0146300. doi: 10.1371/journal.pone.0146300. eCollection 2016. PLoS One. 2016. PMID: 26730820 Free PMC article.
-
biotoolsSchema: a formalized schema for bioinformatics software description.Gigascience. 2021 Jan 27;10(1):giaa157. doi: 10.1093/gigascience/giaa157. Gigascience. 2021. PMID: 33506265 Free PMC article.
-
One Thousand and One Software for Proteomics: Tales of the Toolmakers of Science.J Proteome Res. 2019 Oct 4;18(10):3580-3585. doi: 10.1021/acs.jproteome.9b00219. Epub 2019 Aug 29. J Proteome Res. 2019. PMID: 31429284 Review.
-
Bioinformatics software resources.Brief Bioinform. 2004 Sep;5(3):300-4. doi: 10.1093/bib/5.3.300. Brief Bioinform. 2004. PMID: 15383216 Review.
Cited by
-
Enabling One Health solutions through genomics.Indian J Med Res. 2021 Mar;153(3):273-279. doi: 10.4103/ijmr.IJMR_576_21. Indian J Med Res. 2021. PMID: 33906989 Free PMC article. No abstract available.
-
Implementing the FAIR Data Principles in precision oncology: review of supporting initiatives.Brief Bioinform. 2020 May 21;21(3):936-945. doi: 10.1093/bib/bbz044. Brief Bioinform. 2020. PMID: 31263868 Free PMC article. Review.
-
The role of metadata in reproducible computational research.Patterns (N Y). 2021 Sep 10;2(9):100322. doi: 10.1016/j.patter.2021.100322. eCollection 2021 Sep 10. Patterns (N Y). 2021. PMID: 34553169 Free PMC article. Review.
-
ELIXIR-UK role in bioinformatics training at the national level and across ELIXIR.F1000Res. 2017 Jun 21;6:ELIXIR-952. doi: 10.12688/f1000research.11837.1. eCollection 2017. F1000Res. 2017. PMID: 28781748 Free PMC article.
-
Improving bioinformatics software quality through incorporation of software engineering practices.PeerJ Comput Sci. 2022 Jan 5;8:e839. doi: 10.7717/peerj-cs.839. eCollection 2022. PeerJ Comput Sci. 2022. PMID: 35111923 Free PMC article.
References
-
- May M. Life science technologies: Big biological impacts from big data. Science. 2014;344:1298–1300.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

