Referenced Single-Molecule Measurements Differentiate between GPCR Oligomerization States
- PMID: 26536257
- PMCID: PMC4643199
- DOI: 10.1016/j.bpj.2015.09.004
Referenced Single-Molecule Measurements Differentiate between GPCR Oligomerization States
Abstract
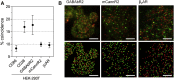
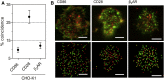
The extent to which Rhodopsin family G-protein-coupled receptors (GPCRs) form invariant oligomers is contentious. Recent single-molecule fluorescence imaging studies mostly argue against the existence of constitutive receptor dimers and instead suggest that GPCRs only dimerize transiently, if at all. However, whether or not even transient dimers exist is not always clear due to difficulties in unambiguously distinguishing genuine interactions from chance colocalizations, particularly with respect to short-lived events. Previous single-molecule studies have depended critically on calculations of chance colocalization rates and/or comparison with unfixed control proteins whose diffusional behavior may or may not differ from that of the test receptor. Here, we describe a single-molecule imaging assay that 1) utilizes comparisons with well-characterized control proteins, i.e., the monomer CD86 and the homodimer CD28, and 2) relies on cell fixation to limit artifacts arising from differences in the distribution and diffusion of test proteins versus these controls. The improved assay reliably reports the stoichiometry of the Glutamate-family GPCR dimer, γ-amino butyric acid receptor b2, whereas two Rhodopsin-family GPCRs, β2-adrenergic receptor and mCannR2, exhibit colocalization levels comparable to those of CD86 monomers, strengthening the case against invariant GPCR oligomerization.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Fluorescence correlation spectroscopy and photon-counting histogram analysis of receptor-receptor interactions.Methods Cell Biol. 2013;117:181-96. doi: 10.1016/B978-0-12-408143-7.00010-4. Methods Cell Biol. 2013. PMID: 24143978
-
Analysis of receptor oligomerization by FRAP microscopy.Nat Methods. 2009 Mar;6(3):225-30. doi: 10.1038/nmeth.1304. Epub 2009 Feb 22. Nat Methods. 2009. PMID: 19234451
-
Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization.Proc Natl Acad Sci U S A. 2013 Jan 8;110(2):743-8. doi: 10.1073/pnas.1205798110. Epub 2012 Dec 24. Proc Natl Acad Sci U S A. 2013. PMID: 23267088 Free PMC article.
-
Revealing G-protein-coupled receptor oligomerization at the single-molecule level through a nanoscopic lens: methods, dynamics and biological function.FEBS J. 2016 Apr;283(7):1197-217. doi: 10.1111/febs.13577. Epub 2015 Nov 28. FEBS J. 2016. PMID: 26509747 Review.
-
GPCR homo-oligomerization.Curr Opin Cell Biol. 2019 Apr;57:40-47. doi: 10.1016/j.ceb.2018.10.007. Epub 2018 Nov 16. Curr Opin Cell Biol. 2019. PMID: 30453145 Free PMC article. Review.
Cited by
-
Diffusion and Oligomerization States of the Muscarinic M1 Receptor in Live Cells─The Impact of Ligands and Membrane Disruptors.J Phys Chem B. 2024 May 9;128(18):4354-4366. doi: 10.1021/acs.jpcb.4c01035. Epub 2024 Apr 29. J Phys Chem B. 2024. PMID: 38683784
-
Receptor Quaternary Organization Explains G Protein-Coupled Receptor Family Structure.Cell Rep. 2017 Sep 12;20(11):2654-2665. doi: 10.1016/j.celrep.2017.08.072. Cell Rep. 2017. PMID: 28903045 Free PMC article.
-
The Class-A GPCR Dopamine D2 Receptor Forms Transient Dimers Stabilized by Agonists: Detection by Single-Molecule Tracking.Cell Biochem Biophys. 2018 Jun;76(1-2):29-37. doi: 10.1007/s12013-017-0829-y. Epub 2017 Nov 7. Cell Biochem Biophys. 2018. PMID: 29116599 Free PMC article.
-
ProDOL: a general method to determine the degree of labeling for staining optimization and molecular counting.Nat Methods. 2024 Sep;21(9):1708-1715. doi: 10.1038/s41592-024-02376-6. Epub 2024 Aug 8. Nat Methods. 2024. PMID: 39117875 Free PMC article.
-
Quantification of absolute labeling efficiency at the single-protein level.Nat Methods. 2024 Sep;21(9):1702-1707. doi: 10.1038/s41592-024-02242-5. Epub 2024 Apr 24. Nat Methods. 2024. PMID: 38658647 Free PMC article.
References
-
- Leake M.C., Chandler J.H., Armitage J.P. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. - PubMed
-
- Kohout S.C., Ulbrich M.H., Isacoff E.Y. Subunit organization and functional transitions in Ci-VSP. Nat. Struct. Mol. Biol. 2008;15:106–108. - PubMed
-
- Moertelmaier M., Brameshuber M., Stockinger H. Thinning out clusters while conserving stoichiometry of labeling. Appl. Phys. Lett. 2005;87:263903.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

