RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics
- PMID: 26525104
- PMCID: PMC4644263
- DOI: 10.1016/j.ccell.2015.09.018
RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics
Abstract
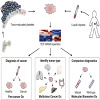
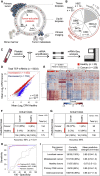
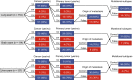
Tumor-educated blood platelets (TEPs) are implicated as central players in the systemic and local responses to tumor growth, thereby altering their RNA profile. We determined the diagnostic potential of TEPs by mRNA sequencing of 283 platelet samples. We distinguished 228 patients with localized and metastasized tumors from 55 healthy individuals with 96% accuracy. Across six different tumor types, the location of the primary tumor was correctly identified with 71% accuracy. Also, MET or HER2-positive, and mutant KRAS, EGFR, or PIK3CA tumors were accurately distinguished using surrogate TEP mRNA profiles. Our results indicate that blood platelets provide a valuable platform for pan-cancer, multiclass cancer, and companion diagnostics, possibly enabling clinical advances in blood-based "liquid biopsies".
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Comment in
-
Cancer genetics: RNA-seq for blood-based pan-cancer diagnostics.Nat Rev Genet. 2015 Dec;16(12):688. doi: 10.1038/nrg4048. Epub 2015 Nov 10. Nat Rev Genet. 2015. PMID: 26553328 No abstract available.
-
Tumor-Educated Platelets as Liquid Biopsy in Cancer Patients.Cancer Cell. 2015 Nov 9;28(5):552-554. doi: 10.1016/j.ccell.2015.10.007. Cancer Cell. 2015. PMID: 26555171
-
Diagnosis: RNA-seq for blood-based pan-cancer diagnostics.Nat Rev Cancer. 2015 Dec;15(12):696-7. doi: 10.1038/nrc4048. Epub 2015 Nov 13. Nat Rev Cancer. 2015. PMID: 26563465 No abstract available.
-
A Word of Caution on New and Revolutionary Diagnostic Tests.Cancer Cell. 2016 Feb 8;29(2):141-2. doi: 10.1016/j.ccell.2016.01.003. Cancer Cell. 2016. PMID: 26859453 No abstract available.
-
Re: a Word of Caution on New and Revolutionary Diagnostic Tests.Cancer Cell. 2016 Feb 8;29(2):143-4. doi: 10.1016/j.ccell.2016.01.004. Cancer Cell. 2016. PMID: 26859454 No abstract available.
Similar articles
-
Construction of a reference material panel for detecting KRAS/NRAS/EGFR/BRAF/MET mutations in plasma ctDNA.J Clin Pathol. 2021 May;74(5):314-320. doi: 10.1136/jclinpath-2020-206745. Epub 2020 Aug 17. J Clin Pathol. 2021. PMID: 32817175 Free PMC article.
-
Concurrent oncogene mutation profile in Chinese patients with stage Ib lung adenocarcinoma.Medicine (Baltimore). 2014 Dec;93(29):e296. doi: 10.1097/MD.0000000000000296. Medicine (Baltimore). 2014. PMID: 25546673 Free PMC article.
-
Clinicopathologic features and outcomes of patients with lung adenocarcinomas harboring BRAF mutations in the Lung Cancer Mutation Consortium.Cancer. 2015 Feb 1;121(3):448-56. doi: 10.1002/cncr.29042. Epub 2014 Oct 1. Cancer. 2015. PMID: 25273224 Free PMC article.
-
Tumor-Educated Platelets as a Noninvasive Biomarker Source for Cancer Detection and Progression Monitoring.Cancer Res. 2018 Jul 1;78(13):3407-3412. doi: 10.1158/0008-5472.CAN-18-0887. Epub 2018 Jun 19. Cancer Res. 2018. PMID: 29921699 Review.
-
Recommendations from the EGAPP Working Group: can testing of tumor tissue for mutations in EGFR pathway downstream effector genes in patients with metastatic colorectal cancer improve health outcomes by guiding decisions regarding anti-EGFR therapy?Genet Med. 2013 Jul;15(7):517-27. doi: 10.1038/gim.2012.184. Epub 2013 Feb 21. Genet Med. 2013. PMID: 23429431
Cited by
-
TRIM27 revealing by tumor educated platelet RNA-sequencing, as a potential biomarker for malignant ground-glass opacities diagnosis mediates glycolysis of non-small cell lung cancer cells partially through HOXM1.Transl Lung Cancer Res. 2024 Sep 30;13(9):2307-2325. doi: 10.21037/tlcr-24-157. Epub 2024 Sep 24. Transl Lung Cancer Res. 2024. PMID: 39430321 Free PMC article.
-
Platelets: The Emerging Clinical Diagnostics and Therapy Selection of Cancer Liquid Biopsies.Onco Targets Ther. 2021 May 25;14:3417-3428. doi: 10.2147/OTT.S311907. eCollection 2021. Onco Targets Ther. 2021. PMID: 34079287 Free PMC article. Review.
-
Immunotherapy in glioblastoma: emerging options in precision medicine.CNS Oncol. 2016 Jul;5(3):175-86. doi: 10.2217/cns-2016-0009. Epub 2016 May 26. CNS Oncol. 2016. PMID: 27225028 Free PMC article. Review.
-
A Comprehensive Review of the Potential Role of Liquid Biopsy as a Diagnostic, Prognostic, and Predictive Biomarker in Pancreatic Ductal Adenocarcinoma.Cells. 2023 Dec 19;13(1):3. doi: 10.3390/cells13010003. Cells. 2023. PMID: 38201207 Free PMC article. Review.
-
Liquid Biopsy in Glioblastoma Management: From Current Research to Future Perspectives.Oncologist. 2021 Oct;26(10):865-878. doi: 10.1002/onco.13858. Epub 2021 Jun 23. Oncologist. 2021. PMID: 34105205 Free PMC article. Review.
References
-
- Alix-Panabières C., Pantel K. Challenges in circulating tumour cell research. Nat. Rev. Cancer. 2014;14:623–631. - PubMed
-
- Alix-Panabières C., Schwarzenbach H., Pantel K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 2012;63:199–215. - PubMed
-
- Bidard F.-C., Peeters D.J., Fehm T., Nolé F., Gisbert-Criado R., Mavroudis D., Grisanti S., Generali D., Garcia-Saenz J.A., Stebbing J. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15:406–414. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

