NCG 5.0: updates of a manually curated repository of cancer genes and associated properties from cancer mutational screenings
- PMID: 26516186
- PMCID: PMC4702816
- DOI: 10.1093/nar/gkv1123
NCG 5.0: updates of a manually curated repository of cancer genes and associated properties from cancer mutational screenings
Abstract
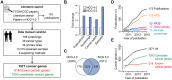
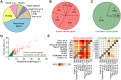
The Network of Cancer Genes (NCG, http://ncg.kcl.ac.uk/) is a manually curated repository of cancer genes derived from the scientific literature. Due to the increasing amount of cancer genomic data, we have introduced a more robust procedure to extract cancer genes from published cancer mutational screenings and two curators independently reviewed each publication. NCG release 5.0 (August 2015) collects 1571 cancer genes from 175 published studies that describe 188 mutational screenings of 13 315 cancer samples from 49 cancer types and 24 primary sites. In addition to collecting cancer genes, NCG also provides information on the experimental validation that supports the role of these genes in cancer and annotates their properties (duplicability, evolutionary origin, expression profile, function and interactions with proteins and miRNAs).
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
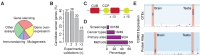

Similar articles
-
The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens.Genome Biol. 2019 Jan 3;20(1):1. doi: 10.1186/s13059-018-1612-0. Genome Biol. 2019. PMID: 30606230 Free PMC article.
-
Network of Cancer Genes: a web resource to analyze duplicability, orthology and network properties of cancer genes.Nucleic Acids Res. 2010 Jan;38(Database issue):D670-5. doi: 10.1093/nar/gkp957. Epub 2009 Nov 11. Nucleic Acids Res. 2010. PMID: 19906700 Free PMC article.
-
NCG 4.0: the network of cancer genes in the era of massive mutational screenings of cancer genomes.Database (Oxford). 2014 Mar 7;2014:bau015. doi: 10.1093/database/bau015. Print 2014. Database (Oxford). 2014. PMID: 24608173 Free PMC article.
-
Low duplicability and network fragility of cancer genes.Trends Genet. 2008 Sep;24(9):427-30. doi: 10.1016/j.tig.2008.06.003. Epub 2008 Jul 31. Trends Genet. 2008. PMID: 18675489 Review.
-
Locus-specific databases in cancer: what future in a post-genomic era? The TP53 LSDB paradigm.Hum Mutat. 2014 Jun;35(6):643-53. doi: 10.1002/humu.22518. Epub 2014 Mar 5. Hum Mutat. 2014. PMID: 24478183 Review.
Cited by
-
Computational approaches for the identification of cancer genes and pathways.Wiley Interdiscip Rev Syst Biol Med. 2017 Jan;9(1):e1364. doi: 10.1002/wsbm.1364. Epub 2016 Nov 11. Wiley Interdiscip Rev Syst Biol Med. 2017. PMID: 27863091 Free PMC article. Review.
-
Prognostic significance of alpha-2-macrglobulin and low-density lipoprotein receptor-related protein-1 in various cancers.Am J Cancer Res. 2024 Jun 15;14(6):3036-3058. doi: 10.62347/VUJV9180. eCollection 2024. Am J Cancer Res. 2024. PMID: 39005669 Free PMC article.
-
Oncodomains: A protein domain-centric framework for analyzing rare variants in tumor samples.PLoS Comput Biol. 2017 Apr 20;13(4):e1005428. doi: 10.1371/journal.pcbi.1005428. eCollection 2017 Apr. PLoS Comput Biol. 2017. PMID: 28426665 Free PMC article.
-
Network analysis identifies common genes associated with obesity in six obesity-related diseases.J Zhejiang Univ Sci B. 2017 Aug.;18(8):727-732. doi: 10.1631/jzus.B1600454. J Zhejiang Univ Sci B. 2017. PMID: 28786249 Free PMC article.
-
Nongenic cancer-risk SNPs affect oncogenes, tumour-suppressor genes, and immune function.Br J Cancer. 2020 Feb;122(4):569-577. doi: 10.1038/s41416-019-0614-3. Epub 2019 Dec 6. Br J Cancer. 2020. PMID: 31806877 Free PMC article.
References
-
- Garraway L.A., Lander E.S. Lessons from the cancer genome. Cell. 2013;153:17–37. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

