Comparing the Assembly and Handedness Dynamics of (H3.3-H4)2 Tetrasomes to Canonical Tetrasomes
- PMID: 26506534
- PMCID: PMC4623960
- DOI: 10.1371/journal.pone.0141267
Comparing the Assembly and Handedness Dynamics of (H3.3-H4)2 Tetrasomes to Canonical Tetrasomes
Abstract
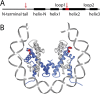
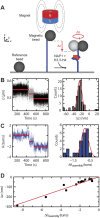
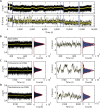
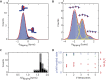
Eukaryotic nucleosomes consists of an (H3-H4)2 tetramer and two H2A-H2B dimers, around which 147 bp of DNA are wrapped in 1.7 left-handed helical turns. During chromatin assembly, the (H3-H4)2 tetramer binds first, forming a tetrasome that likely constitutes an important intermediate during ongoing transcription. We recently showed that (H3-H4)2 tetrasomes spontaneously switch between a left- and right-handed wrapped state of the DNA, a phenomenon that may serve to buffer changes in DNA torque induced by RNA polymerase in transcription. Within nucleosomes of actively transcribed genes, however, canonical H3 is progressively replaced by its variant H3.3. Consequently, one may ask if and how the DNA chirality dynamics of tetrasomes is altered by H3.3. Recent findings that H3.3-containing nucleosomes result in less stable and less condensed chromatin further underline the need to study the microscopic underpinnings of H3.3-containing tetrasomes and nucleosomes. Here we report real-time single-molecule studies of (H3.3-H4)2 tetrasome dynamics using Freely Orbiting Magnetic Tweezers and Electromagnetic Torque Tweezers. We find that the assembly of H3.3-containing tetrasomes and nucleosomes by the histone chaperone Nucleosome Assembly Protein 1 (NAP1) occurs in an identical manner to that of H3-containing tetrasomes and nucleosomes. Likewise, the flipping behavior of DNA handedness in tetrasomes is not impacted by the presence of H3.3. We also examine the effect of free NAP1, H3.3, and H4 in solution on flipping behavior and conclude that the probability for a tetrasome to occupy the left-handed state is only slightly enhanced by the presence of free protein. These data demonstrate that the incorporation of H3.3 does not alter the structural dynamics of tetrasomes, and hence that the preferred incorporation of this histone variant in transcriptionally active regions does not result from its enhanced ability to accommodate torsional stress, but rather may be linked to specific chaperone or remodeler requirements or communication with the nuclear environment.
Conflict of interest statement
Figures
Similar articles
-
Nucleosome assembly dynamics involve spontaneous fluctuations in the handedness of tetrasomes.Cell Rep. 2015 Jan 13;10(2):216-25. doi: 10.1016/j.celrep.2014.12.022. Epub 2015 Jan 8. Cell Rep. 2015. PMID: 25578730
-
Modification of the histone tetramer at the H3-H3 interface impacts tetrasome conformations and dynamics.J Chem Phys. 2018 Mar 28;148(12):123323. doi: 10.1063/1.5009100. J Chem Phys. 2018. PMID: 29604863
-
Nucleosome dynamics. Protein and DNA contributions in the chiral transition of the tetrasome, the histone (H3-H4)2 tetramer-DNA particle.J Mol Biol. 1999 Aug 27;291(4):815-41. doi: 10.1006/jmbi.1999.2988. J Mol Biol. 1999. PMID: 10452891
-
Current progress on structural studies of nucleosomes containing histone H3 variants.Curr Opin Struct Biol. 2013 Feb;23(1):109-15. doi: 10.1016/j.sbi.2012.10.009. Epub 2012 Dec 22. Curr Opin Struct Biol. 2013. PMID: 23265997 Review.
-
Histone dynamics during transcription: exchange of H2A/H2B dimers and H3/H4 tetramers during pol II elongation.Results Probl Cell Differ. 2006;41:77-90. doi: 10.1007/400_009. Results Probl Cell Differ. 2006. PMID: 16909891 Review.
Cited by
-
The Oligomerization Landscape of Histones.Biophys J. 2019 May 21;116(10):1845-1855. doi: 10.1016/j.bpj.2019.03.021. Epub 2019 Apr 17. Biophys J. 2019. PMID: 31005236 Free PMC article.
-
Chiral Systems Made from DNA.Adv Sci (Weinh). 2021 Jan 21;8(5):2003113. doi: 10.1002/advs.202003113. eCollection 2021 Mar. Adv Sci (Weinh). 2021. PMID: 33717850 Free PMC article. Review.
-
The dynamic interplay between DNA topoisomerases and DNA topology.Biophys Rev. 2016 Nov;8(Suppl 1):101-111. doi: 10.1007/s12551-016-0240-8. Epub 2016 Nov 14. Biophys Rev. 2016. PMID: 28510219 Free PMC article. Review.
-
Constructing arrays of nucleosome positioning sequences using Gibson Assembly for single-molecule studies.Sci Rep. 2020 Jun 18;10(1):9903. doi: 10.1038/s41598-020-66259-4. Sci Rep. 2020. PMID: 32555215 Free PMC article.
-
Unravelling DNA Organization with Single-Molecule Force Spectroscopy Using Magnetic Tweezers.Methods Mol Biol. 2024;2819:535-572. doi: 10.1007/978-1-0716-3930-6_25. Methods Mol Biol. 2024. PMID: 39028523
References
-
- Luger K. Structure and dynamic behavior of nucleosomes. Current opinion in genetics & development. 2003;13(2):127–35. . - PubMed
-
- Daban JR, Cantor CR. Structural and kinetic study of the self-assembly of nucleosome core particles. Journal of molecular biology. 1982;156(4):749–69. . - PubMed
-
- Hamiche A, Carot V, Alilat M, De Lucia F, O'Donohue MF, Revet B, et al. Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: Potential flipping of the protein from a left- to a right-handed superhelical form. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(15):7588–93. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

