Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae
- PMID: 26502892
- PMCID: PMC4631204
- DOI: 10.15252/msb.156513
Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae
Figures
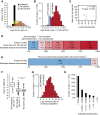
The majority of yeast acetylation sites are highly sensitive to partial chemical acetylation by AcP. The histogram shows the distribution of SILAC L/H ratios for the indicated samples.
AcP caused substantially increased acetylation at a majority of sites. The histogram shows acetylation changes induced by 100 mM AcP in two experimental replicates, and only sites that were independently identified in cells without AcP treatment are shown.
Acetylation stoichiometry is inversely proportional to AcP sensitivity. The scatterplot shows the relationship between AcP sensitivity (SILAC ratio L/H 100 mM AcP) and acetylation stoichiometry (Log10 stoichiometry) as determined by AQUA analysis (Table 1). The Spearman's correlation (ρ) and the significance by two-tailed test (P-value) are shown.
Most acetylation occurs with a low stoichiometry. Absolute acetylation site stoichiometries were estimated based on relative abundance changes (SILAC ratio L/H) after treatment with 100 mM AcP and using an estimate that AcP treatment caused < 1% chemical acetylation.
For comparison, previously determined phosphorylation site stoichiometries are shown *(Wu et al, 2011).
iBAQ-based abundance corrected acetylated peptide intensity (I/iBAQ-A) is proportional to AcP sensitivity. The box plots show the distributions of I/iBAQ-A values for the indicated classes of acetylation sites. AcP-insensitive sites had a significantly (P) higher distribution of I/iBAQ-A values by Wilcoxon test.
Sites without SILAC ratios are highly sensitive to AcP. The minimum ratio of increased acetylation was determined by calculating the increased intensity of AcP-treated “light” peptides relative to an empirically determined detection limit for “heavy” SILAC peptides.
Absolute acetylation stoichiometry of sites without SILAC ratios was estimated to be very low. Stoichiometry was estimated by the same method used to estimate stoichiometry of sites with SILAC ratios in (D) using the minimum ratios of increased acetylation shown in (G).
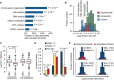
AcP-insensitive acetylation sites occur on proteins associated with nuclear processes. Gene Ontology (GO) term enrichment was performed by comparing proteins with AcP-insensitive acetylation sites (ratio L/H < 2) to all acetylated proteins. The bar graph shows the percentage of AcP-insensitive sites occurring on proteins associated with the indicated GO terms. P-values (P) indicate the statistical significance of GO term enrichment by Fishers exact test.
AcP-insensitive acetylation sites occur on nuclear proteins. The histogram shows the distribution of SILAC L/H ratios occurring on proteins localized to the indicated subcellular compartments.
Acetylated peptides from mitochondrial proteins have a significantly higher median I/iBAQ-A value compared to acetylated peptides from cytoplasmic proteins. The box plots show the iBAQ-based abundance-corrected peptide intensity (I/iBAQ-A) distributions for the indicated classes of peptides occurring on mitochondrial (Mito.) or cytoplasmic (Cyto.) proteins. Significance (P) was determined by Wilcoxon test.
AcP-insensitive sites (SILAC Ratio < 2) have a significantly different subcellular distribution. Sites without SILAC ratios (No ratio) have a similar subcellular distribution to AcP-sensitive sites (SILAC Ratio > 2). The bar graph shows the fraction of sites localized to the indicated subcellular compartments: mitochondria (Mito.), cytoplasm (Cyto.), or nucleus (Nuc.). Significance (P) was calculated by Fisher exact test.
Detection of acetylation sites is biased to occur on abundant proteins, and this bias is more pronounced for sites with the lowest estimated stoichiometries. The histograms show the distributions of iBAQ protein abundances for observed proteins (n = 3,104). The distributions of the indicated classes of acetylation sites occurring exclusively in the indicated subcellular compartments are shown in red. The numbers of acetylated proteins are shown in parenthesis, and the median Log10 iBAQ abundance for these acetylated proteins is shown.
Erratum for
-
Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae.Mol Syst Biol. 2014 Jan 30;10(1):716. doi: 10.1002/msb.134766. Print 2014. Mol Syst Biol. 2014. PMID: 24489116 Free PMC article.
Similar articles
-
Bioinformatics analysis of a Saccharomyces cerevisiae N-terminal proteome provides evidence of alternative translation initiation and post-translational N-terminal acetylation.J Proteome Res. 2011 Aug 5;10(8):3578-89. doi: 10.1021/pr2002325. Epub 2011 Jun 20. J Proteome Res. 2011. PMID: 21619078
-
The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae.Genes Dev. 2001 Dec 1;15(23):3169-82. doi: 10.1101/gad.929001. Genes Dev. 2001. PMID: 11731480 Free PMC article.
-
Insights into the impact of histone acetylation and methylation on Sir protein recruitment, spreading, and silencing in Saccharomyces cerevisiae.J Mol Biol. 2008 Sep 12;381(4):826-44. doi: 10.1016/j.jmb.2008.06.059. Epub 2008 Jun 28. J Mol Biol. 2008. PMID: 18619469
-
SIR2 modifies histone H4-K16 acetylation and affects superhelicity in the ARS region of plasmid chromatin in Saccharomyces cerevisiae.Nucleic Acids Res. 2006;34(19):5426-37. doi: 10.1093/nar/gkl678. Epub 2006 Sep 29. Nucleic Acids Res. 2006. PMID: 17012273 Free PMC article.
-
Untargeted tail acetylation of histones in chromatin: lessons from yeast.Biochem Cell Biol. 2009 Feb;87(1):107-16. doi: 10.1139/O08-097. Biochem Cell Biol. 2009. PMID: 19234527 Review.
Cited by
-
Peptide-Based 2-Aminophenylamide Probes for Targeting Endogenous Class I Histone Deacetylase Complexes.Chembiochem. 2019 Dec 13;20(24):3001-3005. doi: 10.1002/cbic.201900339. Epub 2019 Sep 26. Chembiochem. 2019. PMID: 31270913 Free PMC article.
-
Revealing Dynamic Protein Acetylation across Subcellular Compartments.J Proteome Res. 2020 Jun 5;19(6):2404-2418. doi: 10.1021/acs.jproteome.0c00088. Epub 2020 Apr 27. J Proteome Res. 2020. PMID: 32290654 Free PMC article.
-
Utilizing Optimized Tools to Investigate PTM Crosstalk: Identifying Potential PTM Crosstalk of Acetylated Mitochondrial Proteins.Proteomes. 2018 May 22;6(2):24. doi: 10.3390/proteomes6020024. Proteomes. 2018. PMID: 29786648 Free PMC article.
-
HDAC6-mediated acetylation of lipid droplet-binding protein CIDEC regulates fat-induced lipid storage.J Clin Invest. 2017 Apr 3;127(4):1353-1369. doi: 10.1172/JCI85963. Epub 2017 Mar 13. J Clin Invest. 2017. PMID: 28287402 Free PMC article.
-
A Proteomic Approach to Analyze the Aspirin-mediated Lysine Acetylome.Mol Cell Proteomics. 2017 Feb;16(2):310-326. doi: 10.1074/mcp.O116.065219. Epub 2016 Dec 2. Mol Cell Proteomics. 2017. PMID: 27913581 Free PMC article.
References
-
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Science Signaling. 2010;3:ra3. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

