Metabolic Signaling to Chromatin
- PMID: 26492570
- PMCID: PMC5088527
- DOI: 10.1101/cshperspect.a019463
Metabolic Signaling to Chromatin
Abstract
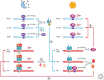
There is a dynamic interplay between metabolic processes and gene regulation via the remodeling of chromatin. Most chromatin-modifying enzymes use cofactors, which are products of metabolic processes. This article explores the biosynthetic pathways of the cofactors nicotinamide adenine dinucleotide (NAD), acetyl coenzyme A (acetyl-CoA), S-adenosyl methionine (SAM), α-ketoglutarate, and flavin adenine dinucleotide (FAD), and their role in metabolically regulating chromatin processes. A more detailed look at the interaction between chromatin and the metabolic processes of circadian rhythms and aging is described as a paradigm for this emerging interdisciplinary field.
Copyright © 2016 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


Similar articles
-
Nutritional Epigenetics: How Metabolism Epigenetically Controls Cellular Physiology, Gene Expression and Disease.Subcell Biochem. 2022;100:239-267. doi: 10.1007/978-3-031-07634-3_8. Subcell Biochem. 2022. PMID: 36301497
-
Chromatin remodeling regulation by small molecules and metabolites.Biochim Biophys Acta. 2010 Oct-Dec;1799(10-12):671-80. doi: 10.1016/j.bbagrm.2010.05.007. Epub 2010 May 20. Biochim Biophys Acta. 2010. PMID: 20493981 Review.
-
Mitochondrial transport and metabolism of the vitamin B-derived cofactors thiamine pyrophosphate, coenzyme A, FAD and NAD+ , and related diseases: A review.IUBMB Life. 2022 Jul;74(7):592-617. doi: 10.1002/iub.2612. Epub 2022 Mar 18. IUBMB Life. 2022. PMID: 35304818 Free PMC article. Review.
-
Metabolic regulation of the plant epigenome.Plant J. 2023 Jun;114(5):1001-1013. doi: 10.1111/tpj.16122. Epub 2023 Feb 10. Plant J. 2023. PMID: 36705504 Review.
-
A regulatory role of NAD redox status on flavin cofactor homeostasis in S. cerevisiae mitochondria.Oxid Med Cell Longev. 2013;2013:612784. doi: 10.1155/2013/612784. Epub 2013 Sep 1. Oxid Med Cell Longev. 2013. PMID: 24078860 Free PMC article.
Cited by
-
Genome Scale-Differential Flux Analysis reveals deregulation of lung cell metabolism on SARS-CoV-2 infection.PLoS Comput Biol. 2021 Apr 9;17(4):e1008860. doi: 10.1371/journal.pcbi.1008860. eCollection 2021 Apr. PLoS Comput Biol. 2021. PMID: 33835998 Free PMC article.
-
The Aging Epigenome.Mol Cell. 2016 Jun 2;62(5):728-44. doi: 10.1016/j.molcel.2016.05.013. Mol Cell. 2016. PMID: 27259204 Free PMC article. Review.
-
Mechanisms contributing to persistently activated cell phenotypes in pulmonary hypertension.J Physiol. 2019 Feb;597(4):1103-1119. doi: 10.1113/JP275857. Epub 2018 Aug 7. J Physiol. 2019. PMID: 29920674 Free PMC article. Review.
-
Epigenetic markers and therapeutic targets for metastasis.Cancer Metastasis Rev. 2023 Jun;42(2):427-443. doi: 10.1007/s10555-023-10109-y. Epub 2023 Jun 7. Cancer Metastasis Rev. 2023. PMID: 37286865 Free PMC article. Review.
-
Epigenome-metabolism nexus in the retina: implications for aging and disease.Trends Genet. 2024 Aug;40(8):718-729. doi: 10.1016/j.tig.2024.04.012. Epub 2024 May 22. Trends Genet. 2024. PMID: 38782642 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
