Mechanistic and Kinetic Differences between Reverse Transcriptases of Vpx Coding and Non-coding Lentiviruses
- PMID: 26483545
- PMCID: PMC4705996
- DOI: 10.1074/jbc.M115.691576
Mechanistic and Kinetic Differences between Reverse Transcriptases of Vpx Coding and Non-coding Lentiviruses
Abstract
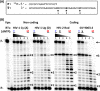
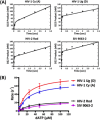
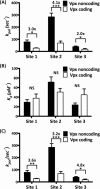
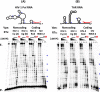
Among lentiviruses, HIV Type 2 (HIV-2) and many simian immunodeficiency virus (SIV) strains replicate rapidly in non-dividing macrophages, whereas HIV Type 1 (HIV-1) replication in this cell type is kinetically delayed. The efficient replication capability of HIV-2/SIV in non-dividing cells is induced by a unique, virally encoded accessory protein, Vpx, which proteasomally degrades the host antiviral restriction factor, SAM domain- and HD domain-containing protein 1 (SAMHD1). SAMHD1 is a dNTPase and kinetically suppresses the reverse transcription step of HIV-1 in macrophages by hydrolyzing and depleting cellular dNTPs. In contrast, Vpx, which is encoded by HIV-2/SIV, kinetically accelerates reverse transcription by counteracting SAMHD1 and then elevating cellular dNTP concentration in non-dividing cells. Here, we conducted the pre-steady-state kinetic analysis of reverse transcriptases (RTs) from two Vpx non-coding and two Vpx coding lentiviruses. At all three sites of the template tested, the two RTs of the Vpx non-coding viruses (HIV-1) displayed higher kpol values than the RTs of the Vpx coding HIV-2/SIV, whereas there was no significant difference in the Kd values of these two groups of RTs. When we employed viral RNA templates that induce RT pausing by their secondary structures, the HIV-1 RTs showed more efficient DNA synthesis through pause sites than the HIV-2/SIV RTs, particularly at low dNTP concentrations found in macrophages. This kinetic study suggests that RTs of the Vpx non-coding HIV-1 may have evolved to execute a faster kpol step, which includes the conformational changes and incorporation chemistry, to counteract the limited dNTP concentration found in non-dividing cells and still promote efficient viral reverse transcription.
Keywords: DNA replication; SAMHD1; Vpx; dNTP; enzyme kinetics; lentivirus; macrophage; reverse transcription.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Similar articles
-
Efficient pre-catalytic conformational change of reverse transcriptases from SAMHD1 non-counteracting primate lentiviruses during dNTP incorporation.Virology. 2019 Nov;537:36-44. doi: 10.1016/j.virol.2019.08.010. Epub 2019 Aug 14. Virology. 2019. PMID: 31442614 Free PMC article.
-
Kinetic variations between reverse transcriptases of viral protein X coding and noncoding lentiviruses.Retrovirology. 2014 Dec 19;11:111. doi: 10.1186/s12977-014-0111-y. Retrovirology. 2014. PMID: 25524560 Free PMC article.
-
Enhanced enzyme kinetics of reverse transcriptase variants cloned from animals infected with SIVmac239 lacking viral protein X.J Biol Chem. 2020 Dec 11;295(50):16975-16986. doi: 10.1074/jbc.RA120.015273. Epub 2020 Oct 2. J Biol Chem. 2020. PMID: 33008888 Free PMC article.
-
Intracellular nucleotide levels and the control of retroviral infections.Virology. 2013 Feb 20;436(2):247-54. doi: 10.1016/j.virol.2012.11.010. Epub 2012 Dec 20. Virology. 2013. PMID: 23260109 Free PMC article. Review.
-
The role of SAMHD1 expression and its relation to HIV-2 (Vpx) gene production.Saudi Pharm J. 2018 Sep;26(6):903-908. doi: 10.1016/j.jsps.2018.03.005. Epub 2018 Mar 12. Saudi Pharm J. 2018. PMID: 30202235 Free PMC article. Review.
Cited by
-
HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins.Proc Natl Acad Sci U S A. 2016 Jul 5;113(27):E3921-30. doi: 10.1073/pnas.1605023113. Epub 2016 Jun 22. Proc Natl Acad Sci U S A. 2016. PMID: 27335459 Free PMC article.
-
Discovery, characterization, and lead optimization of 7-azaindole non-nucleoside HIV-1 reverse transcriptase inhibitors.Bioorg Med Chem Lett. 2016 Aug 15;26(16):4101-5. doi: 10.1016/j.bmcl.2016.06.065. Epub 2016 Jun 25. Bioorg Med Chem Lett. 2016. PMID: 27390064 Free PMC article.
-
Phylogenetic Insights into the Functional Relationship between Primate Lentiviral Reverse Transcriptase and Accessory Proteins Vpx/Vpr.Front Microbiol. 2016 Oct 18;7:1655. doi: 10.3389/fmicb.2016.01655. eCollection 2016. Front Microbiol. 2016. PMID: 27803699 Free PMC article.
-
Cell Cycle Regulation in Macrophages and Susceptibility to HIV-1.Viruses. 2020 Jul 31;12(8):839. doi: 10.3390/v12080839. Viruses. 2020. PMID: 32751972 Free PMC article. Review.
-
Host and Viral Factors Influencing Interplay between the Macrophage and HIV-1.J Neuroimmune Pharmacol. 2019 Mar;14(1):33-43. doi: 10.1007/s11481-018-9795-4. Epub 2018 Jul 11. J Neuroimmune Pharmacol. 2019. PMID: 29995208 Free PMC article. Review.
References
-
- Operario D. J., Reynolds H. M., and Kim B. (2005) Comparison of DNA polymerase activities between recombinant feline immunodeficiency and leukemia virus reverse transcriptases. Virology 335, 106–121 - PubMed
-
- Bjursell G., and Skoog L. (1980) Control of nucleotide pools in mammalian cells. Antibiot. Chemother. 28, 78–85 - PubMed
-
- Cohen A., Barankiewicz J., Lederman H. M., and Gelfand E. W. (1983) Purine and pyrimidine metabolism in human T lymphocytes: regulation of deoxyribonucleotide metabolism. J. Biol. Chem. 258, 12334–12340 - PubMed
-
- Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

