Sleep disruption impairs haematopoietic stem cell transplantation in mice
- PMID: 26465715
- PMCID: PMC4621781
- DOI: 10.1038/ncomms9516
Sleep disruption impairs haematopoietic stem cell transplantation in mice
Abstract
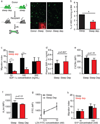
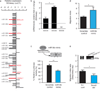
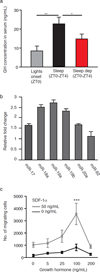
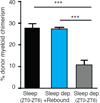
Many of the factors affecting the success of haematopoietic cell transplantation are still unknown. Here we show in mice that donor sleep deprivation reduces the ability of its haematopoietic stem cells (HSCs) to engraft and reconstitute the blood and bone marrow of an irradiated recipient by more than 50%. We demonstrate that sleep deprivation downregulates the expression of microRNA (miR)-19b, a negative regulator of the suppressor of cytokine signalling (SOCS) genes, which inhibit HSC migration and homing. Accordingly, HSCs from sleep-deprived mice have higher levels of SOCS genes expression, lower migration capacity in vitro and reduced homing to the bone marrow in vivo. Recovery of sleep after sleep deprivation restored the reconstitution potential of the HSCs. Taken together, this study provides insights into cellular and molecular mechanisms underlying the effects of sleep deprivation on HSCs, emphasizing the potentially critical role of donor sleep in the success of bone marrow transplantation.
Figures

Similar articles
-
Pyruvate dehydrogenase kinase 1 is essential for transplantable mouse bone marrow hematopoietic stem cell and progenitor function.PLoS One. 2017 Feb 9;12(2):e0171714. doi: 10.1371/journal.pone.0171714. eCollection 2017. PLoS One. 2017. PMID: 28182733 Free PMC article.
-
Evidence that β7 Integrin Regulates Hematopoietic Stem Cell Homing and Engraftment Through Interaction with MAdCAM-1.Stem Cells Dev. 2016 Jan 1;25(1):18-26. doi: 10.1089/scd.2014.0551. Epub 2015 Nov 5. Stem Cells Dev. 2016. PMID: 26422691 Free PMC article.
-
Pre-Transplantation Blockade of TNF-α-Mediated Oxygen Species Accumulation Protects Hematopoietic Stem Cells.Stem Cells. 2017 Apr;35(4):989-1002. doi: 10.1002/stem.2524. Epub 2016 Nov 11. Stem Cells. 2017. PMID: 27753160
-
Analyzing hematopoietic stem cell homing, lodgment, and engraftment to better understand the bone marrow niche.Ann N Y Acad Sci. 2014 Mar;1310:119-28. doi: 10.1111/nyas.12329. Epub 2014 Jan 15. Ann N Y Acad Sci. 2014. PMID: 24428368 Review.
-
Mobilization and homing of hematopoietic stem cells.Adv Exp Med Biol. 2012;741:152-70. doi: 10.1007/978-1-4614-2098-9_11. Adv Exp Med Biol. 2012. PMID: 22457109 Review.
Cited by
-
Systemic and local regulation of hematopoietic homeostasis in health and disease.Nat Cardiovasc Res. 2024 Jun;3(6):651-665. doi: 10.1038/s44161-024-00482-4. Epub 2024 Jun 12. Nat Cardiovasc Res. 2024. PMID: 39196230 Review.
-
Oncogenic role of microRNA-19b-3p-mediated SOCS3 in glioma through activation of JAK-STAT pathway.Metab Brain Dis. 2023 Mar;38(3):945-960. doi: 10.1007/s11011-022-01136-9. Epub 2022 Dec 9. Metab Brain Dis. 2023. PMID: 36484970
-
The contribution of sleep to the neuroendocrine regulation of rhythms in human leukocyte traffic.Semin Immunopathol. 2022 Mar;44(2):239-254. doi: 10.1007/s00281-021-00904-6. Epub 2022 Jan 18. Semin Immunopathol. 2022. PMID: 35041075 Free PMC article. Review.
-
Neuronal regulation of immunity: why, how and where?Nat Rev Immunol. 2021 Jan;21(1):20-36. doi: 10.1038/s41577-020-0387-1. Epub 2020 Aug 18. Nat Rev Immunol. 2021. PMID: 32811994 Review.
-
Single cell RNA sequencing identifies an early monocyte gene signature in acute respiratory distress syndrome.JCI Insight. 2020 Jul 9;5(13):e135678. doi: 10.1172/jci.insight.135678. JCI Insight. 2020. PMID: 32554932 Free PMC article.
References
-
- Bryant PA, Trinder J, Curtis N. Sick and tired: Does sleep have a vital role in the immune system? Nat. Rev. Immunol. 2004;4:457–467. - PubMed
-
- Guariniello LD, Vicari P, Lee KS, de Oliveira AC, Tufik S. Bone marrow and peripheral white blood cells number is affected by sleep deprivation in a murine experimental model. J. Cell. Physiol. 2012;227:361–366. - PubMed
-
- Tsinkalovsky O, et al. Circadian expression of clock genes in purified hematopoietic stem cells is developmentally regulated in mouse bone marrow. Exp. Hematol. 2006;34:1249–1261. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

