Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration
- PMID: 26460020
- PMCID: PMC4629334
- DOI: 10.1073/pnas.1518361112
Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration
Abstract
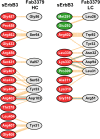
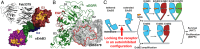
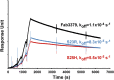
ErbB3 (HER3) is a member of the EGF receptor (EGFR) family of receptor tyrosine kinases, which, unlike the other three family members, contains a pseudo kinase in place of a tyrosine kinase domain. In cancer, ErbB3 activation is driven by a ligand-dependent mechanism through the formation of heterodimers with EGFR, ErbB2, or ErbB4 or via a ligand-independent process through heterodimerization with ErbB2 overexpressed in breast tumors or other cancers. Here we describe the crystal structure of the Fab fragment of an antagonistic monoclonal antibody KTN3379, currently in clinical development in human cancer patients, in complex with the ErbB3 extracellular domain. The structure reveals a unique allosteric mechanism for inhibition of ligand-dependent or ligand-independent ErbB3-driven cancers by binding to an epitope that locks ErbB3 in an inactive conformation. Given the similarities in the mechanism of ErbB receptor family activation, these findings could facilitate structure-based design of antibodies that inhibit EGFR and ErbB4 by an allosteric mechanism.
Keywords: cancer; cell signaling; crystal structure; surface receptor; therapeutic antibodies.
Conflict of interest statement
Conflict of interest statement: J.S. is a founder and consultant of Kolltan. G.F.L., J.S.L., E.J.N., D.A., and Y.H. are Kolltan employees.
Figures
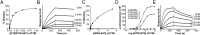
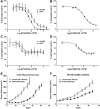
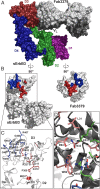

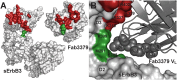

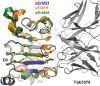
Similar articles
-
A Potent HER3 Monoclonal Antibody That Blocks Both Ligand-Dependent and -Independent Activities: Differential Impacts of PTEN Status on Tumor Response.Mol Cancer Ther. 2016 Apr;15(4):689-701. doi: 10.1158/1535-7163.MCT-15-0555. Epub 2016 Feb 15. Mol Cancer Ther. 2016. PMID: 26880266
-
The ErbB/HER family of protein-tyrosine kinases and cancer.Pharmacol Res. 2014 Jan;79:34-74. doi: 10.1016/j.phrs.2013.11.002. Epub 2013 Nov 20. Pharmacol Res. 2014. PMID: 24269963 Review.
-
Combining anti-ERBB3 antibodies specific for domain I and domain III enhances the anti-tumor activity over the individual monoclonal antibodies.PLoS One. 2014 Nov 11;9(11):e112376. doi: 10.1371/journal.pone.0112376. eCollection 2014. PLoS One. 2014. PMID: 25386657 Free PMC article.
-
Growth stimulation of non-small cell lung cancer cell lines by antibody against epidermal growth factor receptor promoting formation of ErbB2/ErbB3 heterodimers.Mol Cancer Res. 2007 Apr;5(4):393-401. doi: 10.1158/1541-7786.MCR-06-0303. Mol Cancer Res. 2007. PMID: 17426253
-
EGFR family: structure physiology signalling and therapeutic targets.Growth Factors. 2008 Oct;26(5):263-74. doi: 10.1080/08977190802312844. Growth Factors. 2008. PMID: 18800267 Review.
Cited by
-
A small molecule inhibitor of HER3: a proof-of-concept study.Biochem J. 2020 Sep 18;477(17):3329-3347. doi: 10.1042/BCJ20200496. Biochem J. 2020. PMID: 32815546 Free PMC article.
-
A protein network map of head and neck cancer reveals PIK3CA mutant drug sensitivity.Science. 2021 Oct;374(6563):eabf2911. doi: 10.1126/science.abf2911. Epub 2021 Oct 1. Science. 2021. PMID: 34591642 Free PMC article.
-
ERBB3: A potential serum biomarker for early detection and therapeutic target for devil facial tumour 1 (DFT1).PLoS One. 2017 Jun 7;12(6):e0177919. doi: 10.1371/journal.pone.0177919. eCollection 2017. PLoS One. 2017. PMID: 28591206 Free PMC article.
-
Targeting ErbB3 Receptor in Cancer with Inhibitory Antibodies from Llama.Biomedicines. 2021 Aug 28;9(9):1106. doi: 10.3390/biomedicines9091106. Biomedicines. 2021. PMID: 34572289 Free PMC article.
-
Synthesis of Multiple Bispecific Antibody Formats with Only One Single Enzyme Based on Enhanced Trypsiligase.Int J Mol Sci. 2022 Mar 15;23(6):3144. doi: 10.3390/ijms23063144. Int J Mol Sci. 2022. PMID: 35328563 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

