The Interaction of CCDC104/BARTL1 with Arl3 and Implications for Ciliary Function
- PMID: 26455799
- PMCID: PMC4635315
- DOI: 10.1016/j.str.2015.08.016
The Interaction of CCDC104/BARTL1 with Arl3 and Implications for Ciliary Function
Abstract
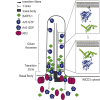
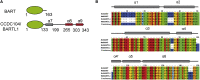
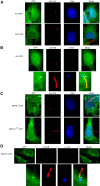
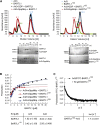
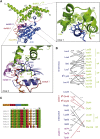
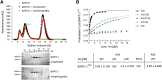
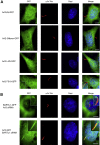
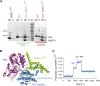
Cilia are small antenna-like cellular protrusions critical for many developmental signaling pathways. The ciliary protein Arl3 has been shown to act as a specific release factor for myristoylated and farnesylated ciliary cargo molecules by binding to the effectors Unc119 and PDE6δ. Here we describe a newly identified Arl3 binding partner, CCDC104/CFAP36. Biochemical and structural analyses reveal that the protein contains a BART-like domain and is called BARTL1. It recognizes an LLxILxxL motif at the N-terminal amphipathic helix of Arl3, which is crucial for the interaction with the BART-like domain but also for the ciliary localization of Arl3 itself. These results seem to suggest a ciliary role of BARTL1, and possibly link it to the Arl3 transport network. We thus speculate on a regulatory mechanism whereby BARTL1 aids the presentation of active Arl3 to its GTPase-activating protein RP2 or hinders Arl3 membrane binding in the area of the transition zone.
Copyright © 2015 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures

Similar articles
-
Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119.EMBO J. 2012 Oct 17;31(20):4085-94. doi: 10.1038/emboj.2012.257. Epub 2012 Sep 7. EMBO J. 2012. PMID: 22960633 Free PMC article.
-
A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins.Elife. 2015 Nov 9;4:e11859. doi: 10.7554/eLife.11859. Elife. 2015. PMID: 26551564 Free PMC article.
-
Arl3 and RP2 regulate the trafficking of ciliary tip kinesins.Hum Mol Genet. 2017 Jul 1;26(13):2480-2492. doi: 10.1093/hmg/ddx143. Hum Mol Genet. 2017. PMID: 28444310 Free PMC article.
-
Sorting of lipidated cargo by the Arl2/Arl3 system.Small GTPases. 2016 Oct;7(4):222-230. doi: 10.1080/21541248.2016.1224454. Epub 2016 Nov 2. Small GTPases. 2016. PMID: 27806215 Free PMC article. Review.
-
Arl3 and RP2 mediated assembly and traffic of membrane associated cilia proteins.Vision Res. 2012 Dec 15;75:2-4. doi: 10.1016/j.visres.2012.07.016. Epub 2012 Aug 2. Vision Res. 2012. PMID: 22884633 Review.
Cited by
-
Novel Biochemical and Structural Insights into the Interaction of Myristoylated Cargo with Unc119 Protein and Their Release by Arl2/3.J Biol Chem. 2016 Sep 23;291(39):20766-78. doi: 10.1074/jbc.M116.741827. Epub 2016 Aug 1. J Biol Chem. 2016. PMID: 27481943 Free PMC article.
-
Allosteric regulation of Arf GTPases and their GEFs at the membrane interface.Small GTPases. 2016 Oct;7(4):283-296. doi: 10.1080/21541248.2016.1215778. Epub 2016 Jul 22. Small GTPases. 2016. PMID: 27449855 Free PMC article. Review.
-
ARL3 mediates BBSome ciliary turnover by promoting its outward movement across the transition zone.J Cell Biol. 2022 Oct 3;221(10):e202111076. doi: 10.1083/jcb.202111076. Epub 2022 Sep 21. J Cell Biol. 2022. PMID: 36129685 Free PMC article.
-
The unusual flagellar-targeting mechanism and functions of the trypanosome ortholog of the ciliary GTPase Arl13b.J Cell Sci. 2018 Sep 5;131(17):jcs219071. doi: 10.1242/jcs.219071. J Cell Sci. 2018. PMID: 30097558 Free PMC article.
-
ARL3 activation requires the co-GEF BART and effector-mediated turnover.Elife. 2021 Jan 13;10:e64624. doi: 10.7554/eLife.64624. Elife. 2021. PMID: 33438581 Free PMC article.
References
-
- Avidor-Reiss T., Maer A.M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C.S. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. - PubMed
-
- Berken A., Thomas C., Wittinghofer A. A new family of RhoGEFs activates the Rop molecular switch in plants. Nature. 2005;436:1176–1180. - PubMed
-
- Chandra A., Grecco H.E., Pisupati V., Perera D., Cassidy L., Skoulidis F., Ismail S.A., Hedberg C., Hanzal-Bayer M., Venkitaraman A.R. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 2012;14:148–158. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

