Long-Range Chromatin Interactions
- PMID: 26430217
- PMCID: PMC4588061
- DOI: 10.1101/cshperspect.a019356
Long-Range Chromatin Interactions
Abstract
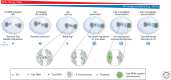
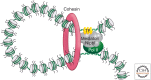
To accommodate genomes in the limited space of the cell nucleus and ensure the correct execution of gene expression programs, genomes are packaged in complex fashion in the three-dimensional cell nucleus. As a consequence of the extensive higher-order organization of chromosomes, distantly located genomic regions on the same or distinct chromosomes undergo long-range interactions. This article discusses the nature of long interactions, mechanisms of their formation, and their emerging functional roles in gene regulation and genome maintenance.
Copyright © 2015 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
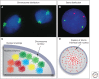
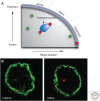

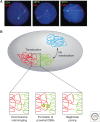
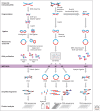

Similar articles
-
The contribution of nuclear compartmentalization to gene regulation.Cell. 2002 Feb 22;108(4):513-21. doi: 10.1016/s0092-8674(02)00650-5. Cell. 2002. PMID: 11909522 Review.
-
The cell nucleus taking centre stage. Workshop on the functional organization of the cell nucleus.EMBO Rep. 2006 Dec;7(12):1211-5. doi: 10.1038/sj.embor.7400840. Epub 2006 Oct 27. EMBO Rep. 2006. PMID: 17068488 Free PMC article. No abstract available.
-
Chromatin organization in the mammalian nucleus.Int Rev Cytol. 2005;242:283-336. doi: 10.1016/S0074-7696(04)42007-5. Int Rev Cytol. 2005. PMID: 15598472 Review.
-
The budding yeast nucleus.Cold Spring Harb Perspect Biol. 2010 Aug;2(8):a000612. doi: 10.1101/cshperspect.a000612. Epub 2010 Jun 16. Cold Spring Harb Perspect Biol. 2010. PMID: 20554704 Free PMC article. Review.
-
Functional architecture in the cell nucleus.Biochem J. 2001 Jun 1;356(Pt 2):297-310. doi: 10.1042/0264-6021:3560297. Biochem J. 2001. PMID: 11368755 Free PMC article. Review.
Cited by
-
Cohesin prevents cross-domain gene coactivation.Nat Genet. 2024 Aug;56(8):1654-1664. doi: 10.1038/s41588-024-01852-1. Epub 2024 Jul 24. Nat Genet. 2024. PMID: 39048795 Free PMC article.
-
The Self-Organizing Genome: Principles of Genome Architecture and Function.Cell. 2020 Oct 1;183(1):28-45. doi: 10.1016/j.cell.2020.09.014. Epub 2020 Sep 24. Cell. 2020. PMID: 32976797 Free PMC article. Review.
-
LDB1 establishes multi-enhancer networks to regulate gene expression.bioRxiv [Preprint]. 2024 Aug 24:2024.08.23.609430. doi: 10.1101/2024.08.23.609430. bioRxiv. 2024. Update in: Mol Cell. 2025 Jan 16;85(2):376-393.e9. doi: 10.1016/j.molcel.2024.11.037. PMID: 39229045 Free PMC article. Updated. Preprint.
-
Regulating Methylation at H3K27: A Trick or Treat for Cancer Cell Plasticity.Cancers (Basel). 2020 Sep 29;12(10):2792. doi: 10.3390/cancers12102792. Cancers (Basel). 2020. PMID: 33003334 Free PMC article. Review.
-
Programmed DNA elimination in the parasitic nematode Ascaris.PLoS Pathog. 2023 Feb 2;19(2):e1011087. doi: 10.1371/journal.ppat.1011087. eCollection 2023 Feb. PLoS Pathog. 2023. PMID: 36730159 Free PMC article. Review.
References
-
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. 2007. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science 318: 1632–1636. - PubMed
-
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol 8: 293–299. - PubMed
-
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. 2011. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144: 214–226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
