SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef
- PMID: 26416733
- PMCID: PMC4600458
- DOI: 10.1038/nature15400
SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef
Abstract
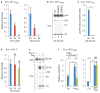
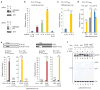
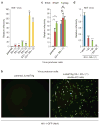
HIV-1 Nef and the unrelated mouse leukaemia virus glycosylated Gag (glycoGag) strongly enhance the infectivity of HIV-1 virions produced in certain cell types in a clathrin-dependent manner. Here we show that Nef and glycoGag prevent the incorporation of the multipass transmembrane proteins serine incorporator 3 (SERINC3) and SERINC5 into HIV-1 virions to an extent that correlates with infectivity enhancement. Silencing of both SERINC3 and SERINC5 precisely phenocopied the effects of Nef and glycoGag on HIV-1 infectivity. The infectivity of nef-deficient virions increased more than 100-fold when produced in double-knockout human CD4(+) T cells that lack both SERINC3 and SERINC5, and re-expression experiments confirmed that the absence of SERINC3 and SERINC5 accounted for the infectivity enhancement. Furthermore, SERINC3 and SERINC5 together restricted HIV-1 replication, and this restriction was evaded by Nef. SERINC3 and SERINC5 are highly expressed in primary human HIV-1 target cells, and inhibiting their downregulation by Nef is a potential strategy to combat HIV/AIDS.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


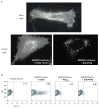
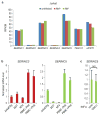
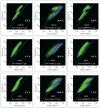
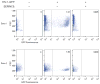




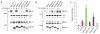
Comment in
-
HIV: Antiviral action countered by Nef.Nature. 2015 Oct 8;526(7572):202-3. doi: 10.1038/nature15637. Epub 2015 Sep 30. Nature. 2015. PMID: 26416750 No abstract available.
-
Viral pathogenesis: HIV-1 Nef targets restriction factors.Nat Rev Microbiol. 2015 Nov;13(11):660-1. doi: 10.1038/nrmicro3573. Epub 2015 Oct 12. Nat Rev Microbiol. 2015. PMID: 26456923 No abstract available.
Similar articles
-
Potent Enhancement of HIV-1 Replication by Nef in the Absence of SERINC3 and SERINC5.mBio. 2019 Jun 11;10(3):e01071-19. doi: 10.1128/mBio.01071-19. mBio. 2019. PMID: 31186327 Free PMC article.
-
HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation.Nature. 2015 Oct 8;526(7572):212-7. doi: 10.1038/nature15399. Epub 2015 Sep 30. Nature. 2015. PMID: 26416734 Free PMC article.
-
Spotlight on HIV-1 Nef: SERINC3 and SERINC5 Identified as Restriction Factors Antagonized by the Pathogenesis Factor.Viruses. 2015 Dec 19;7(12):6730-8. doi: 10.3390/v7122970. Viruses. 2015. PMID: 26703715 Free PMC article.
-
SERINC as a Restriction Factor to Inhibit Viral Infectivity and the Interaction with HIV.J Immunol Res. 2017;2017:1548905. doi: 10.1155/2017/1548905. Epub 2017 Nov 22. J Immunol Res. 2017. PMID: 29359168 Free PMC article. Review.
-
HIV-1 Nef: Taking Control of Protein Trafficking.Traffic. 2016 Sep;17(9):976-96. doi: 10.1111/tra.12412. Epub 2016 Jun 3. Traffic. 2016. PMID: 27161574 Review.
Cited by
-
Prevention and treatment of HIV infection and cognitive disease in mice by innate immune responses.Brain Behav Immun Health. 2020 Mar;3:100054. doi: 10.1016/j.bbih.2020.100054. Epub 2020 Feb 23. Brain Behav Immun Health. 2020. PMID: 32699842 Free PMC article.
-
SARS-CoV-2 ORF7a Mutation Found in BF.5 and BF.7 Sublineages Impacts Its Functions.Int J Mol Sci. 2024 Feb 16;25(4):2351. doi: 10.3390/ijms25042351. Int J Mol Sci. 2024. PMID: 38397027 Free PMC article.
-
SARS-CoV-2-encoded small RNAs are able to repress the host expression of SERINC5 to facilitate viral replication.Front Microbiol. 2023 Feb 16;14:1066493. doi: 10.3389/fmicb.2023.1066493. eCollection 2023. Front Microbiol. 2023. PMID: 36876111 Free PMC article.
-
Two Functional Variants of AP-1 Complexes Composed of either γ2 or γ1 Subunits Are Independently Required for Major Histocompatibility Complex Class I Downregulation by HIV-1 Nef.J Virol. 2020 Mar 17;94(7):e02039-19. doi: 10.1128/JVI.02039-19. Print 2020 Mar 17. J Virol. 2020. PMID: 31915283 Free PMC article.
-
Isolation of Cells from Glioblastoma Multiforme Grade 4 Tumors for Infection with Zika Virus prME and ME Pseudotyped HIV-1.Int J Mol Sci. 2023 Feb 24;24(5):4467. doi: 10.3390/ijms24054467. Int J Mol Sci. 2023. PMID: 36901897 Free PMC article.
References
-
- Kestler HW, 3rd, et al. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–62. - PubMed
-
- Deacon NJ, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

