Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins
- PMID: 26412307
- PMCID: PMC4609299
- DOI: 10.1016/j.molcel.2015.08.018
Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins
Abstract
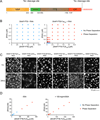
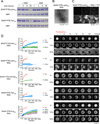
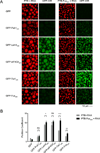
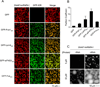
Eukaryotic cells possess numerous dynamic membrane-less organelles, RNP granules, enriched in RNA and RNA-binding proteins containing disordered regions. We demonstrate that the disordered regions of key RNP granule components and the full-length granule protein hnRNPA1 can phase separate in vitro, producing dynamic liquid droplets. Phase separation is promoted by low salt concentrations or RNA. Over time, the droplets mature to more stable states, as assessed by slowed fluorescence recovery after photobleaching and resistance to salt. Maturation often coincides with formation of fibrous structures. Different disordered domains can co-assemble into phase-separated droplets. These biophysical properties demonstrate a plausible mechanism by which interactions between disordered regions, coupled with RNA binding, could contribute to RNP granule assembly in vivo through promoting phase separation. Progression from dynamic liquids to stable fibers may be regulated to produce cellular structures with diverse physiochemical properties and functions. Misregulation could contribute to diseases involving aberrant RNA granules.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II.Mol Cell. 2015 Oct 15;60(2):231-41. doi: 10.1016/j.molcel.2015.09.006. Epub 2015 Oct 8. Mol Cell. 2015. PMID: 26455390 Free PMC article.
-
Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization.Cell. 2015 Sep 24;163(1):123-33. doi: 10.1016/j.cell.2015.09.015. Cell. 2015. PMID: 26406374 Free PMC article.
-
Electrostatic modulation of hnRNPA1 low-complexity domain liquid-liquid phase separation and aggregation.Protein Sci. 2021 Jul;30(7):1408-1417. doi: 10.1002/pro.4108. Epub 2021 May 22. Protein Sci. 2021. PMID: 33982369 Free PMC article.
-
Multiple Modes of Protein-Protein Interactions Promote RNP Granule Assembly.J Mol Biol. 2018 Nov 2;430(23):4636-4649. doi: 10.1016/j.jmb.2018.08.005. Epub 2018 Aug 9. J Mol Biol. 2018. PMID: 30099026 Free PMC article. Review.
-
Membraneless organelles: P granules in Caenorhabditis elegans.Traffic. 2019 Jun;20(6):373-379. doi: 10.1111/tra.12644. Epub 2019 Apr 11. Traffic. 2019. PMID: 30924287 Free PMC article. Review.
Cited by
-
Role and therapeutic potential of liquid-liquid phase separation in amyotrophic lateral sclerosis.J Mol Cell Biol. 2021 Apr 10;13(1):15-28. doi: 10.1093/jmcb/mjaa049. J Mol Cell Biol. 2021. PMID: 32976566 Free PMC article. Review.
-
Long-term memory consolidation: The role of RNA-binding proteins with prion-like domains.RNA Biol. 2017 May 4;14(5):568-586. doi: 10.1080/15476286.2016.1244588. Epub 2016 Oct 11. RNA Biol. 2017. PMID: 27726526 Free PMC article. Review.
-
METTL3-mediated chromatin contacts promote stress granule phase separation through metabolic reprogramming during senescence.Nat Commun. 2024 Jun 26;15(1):5410. doi: 10.1038/s41467-024-49745-5. Nat Commun. 2024. PMID: 38926365 Free PMC article.
-
Atomic resolution map of the solvent interactions driving SOD1 unfolding in CAPRIN1 condensates.Proc Natl Acad Sci U S A. 2024 Aug 27;121(35):e2408554121. doi: 10.1073/pnas.2408554121. Epub 2024 Aug 22. Proc Natl Acad Sci U S A. 2024. PMID: 39172789
-
Rapid and reversible dissolution of biomolecular condensates using light-controlled recruitment of a solubility tag.Nat Commun. 2024 Aug 7;15(1):6717. doi: 10.1038/s41467-024-50858-0. Nat Commun. 2024. PMID: 39112465 Free PMC article.
References
-
- Asherie N, Pande J, Lomakin A, Ogun O, Hanson SR, Smith JB, Benedek GB. Oligomerization and phase separation in globular protein solutions. Biophysical Chemistry. 1998;75:213–227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

