Metabolic plasticity of metastatic breast cancer cells: adaptation to changes in the microenvironment
- PMID: 26408259
- PMCID: PMC4674487
- DOI: 10.1016/j.neo.2015.08.005
Metabolic plasticity of metastatic breast cancer cells: adaptation to changes in the microenvironment
Abstract
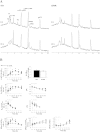
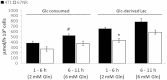
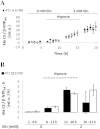
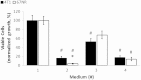
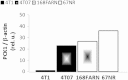
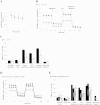
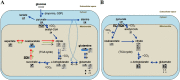
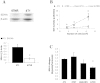
Cancer cells adapt their metabolism during tumorigenesis. We studied two isogenic breast cancer cells lines (highly metastatic 4T1; nonmetastatic 67NR) to identify differences in their glucose and glutamine metabolism in response to metabolic and environmental stress. Dynamic magnetic resonance spectroscopy of (13)C-isotopomers showed that 4T1 cells have higher glycolytic and tricarboxylic acid (TCA) cycle flux than 67NR cells and readily switch between glycolysis and oxidative phosphorylation (OXPHOS) in response to different extracellular environments. OXPHOS activity increased with metastatic potential in isogenic cell lines derived from the same primary breast cancer: 4T1 > 4T07 and 168FARN (local micrometastasis only) > 67NR. We observed a restricted TCA cycle flux at the succinate dehydrogenase step in 67NR cells (but not in 4T1 cells), leading to succinate accumulation and hindering OXPHOS. In the four isogenic cell lines, environmental stresses modulated succinate dehydrogenase subunit A expression according to metastatic potential. Moreover, glucose-derived lactate production was more glutamine dependent in cell lines with higher metastatic potential. These studies show clear differences in TCA cycle metabolism between 4T1 and 67NR breast cancer cells. They indicate that metastases-forming 4T1 cells are more adept at adjusting their metabolism in response to environmental stress than isogenic, nonmetastatic 67NR cells. We suggest that the metabolic plasticity and adaptability are more important to the metastatic breast cancer phenotype than rapid cell proliferation alone, which could 1) provide a new biomarker for early detection of this phenotype, possibly at the time of diagnosis, and 2) lead to new treatment strategies of metastatic breast cancer by targeting mitochondrial metabolism.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







Similar articles
-
Bcl-2/adenovirus E1B 19 kDa interacting protein-3 knockdown enables growth of breast cancer metastases in the lung, liver, and bone.Cancer Res. 2005 Dec 15;65(24):11689-93. doi: 10.1158/0008-5472.CAN-05-3091. Cancer Res. 2005. PMID: 16357180
-
Functional and molecular characterisation of EO771.LMB tumours, a new C57BL/6-mouse-derived model of spontaneously metastatic mammary cancer.Dis Model Mech. 2015 Mar;8(3):237-51. doi: 10.1242/dmm.017830. Epub 2015 Jan 29. Dis Model Mech. 2015. PMID: 25633981 Free PMC article.
-
Metabolic imaging: a link between lactate dehydrogenase A, lactate, and tumor phenotype.Clin Cancer Res. 2011 Oct 1;17(19):6250-6261. doi: 10.1158/1078-0432.CCR-11-0397. Epub 2011 Aug 15. Clin Cancer Res. 2011. PMID: 21844011 Free PMC article.
-
The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism.Cancer Lett. 2015 Jan 28;356(2 Pt A):156-64. doi: 10.1016/j.canlet.2014.04.001. Epub 2014 Apr 13. Cancer Lett. 2015. PMID: 24732809 Free PMC article. Review.
-
Mitochondria Targeting as an Effective Strategy for Cancer Therapy.Int J Mol Sci. 2020 May 9;21(9):3363. doi: 10.3390/ijms21093363. Int J Mol Sci. 2020. PMID: 32397535 Free PMC article. Review.
Cited by
-
Metabolic Reprogramming in Breast Cancer and Its Therapeutic Implications.Cells. 2019 Jan 26;8(2):89. doi: 10.3390/cells8020089. Cells. 2019. PMID: 30691108 Free PMC article. Review.
-
Modulated Electro-Hyperthermia Induces a Prominent Local Stress Response and Growth Inhibition in Mouse Breast Cancer Isografts.Cancers (Basel). 2021 Apr 6;13(7):1744. doi: 10.3390/cancers13071744. Cancers (Basel). 2021. PMID: 33917524 Free PMC article.
-
Shikonin blocks CAF-induced TNBC metastasis by suppressing mitochondrial biogenesis through GSK-3β/NEDD4-1 mediated phosphorylation-dependent degradation of PGC-1α.J Exp Clin Cancer Res. 2024 Jun 27;43(1):180. doi: 10.1186/s13046-024-03101-z. J Exp Clin Cancer Res. 2024. PMID: 38937832 Free PMC article.
-
Prognosis prediction and risk stratification of breast cancer patients based on a mitochondria-related gene signature.Sci Rep. 2024 Feb 3;14(1):2859. doi: 10.1038/s41598-024-52981-w. Sci Rep. 2024. PMID: 38310106 Free PMC article.
-
Metaboloptics: Visualization of the tumor functional landscape via metabolic and vascular imaging.Sci Rep. 2018 Mar 8;8(1):4171. doi: 10.1038/s41598-018-22480-w. Sci Rep. 2018. PMID: 29520098 Free PMC article.
References
-
- Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. - PubMed
-
- Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–895. - PubMed
-
- Bianchini G, Qi Y, Alvarez RH, Iwamoto T, Coutant C, Ibrahim NK, Valero V, Cristofanilli M, Green MC, Radvanyi L. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J Clin Oncol. 2010;28:4316–4323. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

