A high-resolution imaging approach to investigate chromatin architecture in complex tissues
- PMID: 26406379
- PMCID: PMC4583660
- DOI: 10.1016/j.cell.2015.09.002
A high-resolution imaging approach to investigate chromatin architecture in complex tissues
Abstract
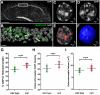
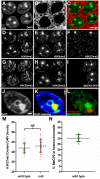
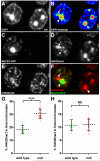
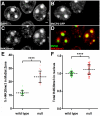
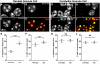
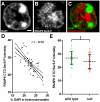
We present ChromATin, a quantitative high-resolution imaging approach for investigating chromatin organization in complex tissues. This method combines analysis of epigenetic modifications by immunostaining, localization of specific DNA sequences by FISH, and high-resolution segregation of nuclear compartments using array tomography (AT) imaging. We then apply this approach to examine how the genome is organized in the mammalian brain using female Rett syndrome mice, which are a mosaic of normal and Mecp2-null cells. Side-by-side comparisons within the same field reveal distinct heterochromatin territories in wild-type neurons that are altered in Mecp2-null nuclei. Mutant neurons exhibit increased chromatin compaction and a striking redistribution of the H4K20me3 histone modification into pericentromeric heterochromatin, a territory occupied normally by MeCP2. These events are not observed in every neuronal cell type, highlighting ChromATin as a powerful in situ method for examining cell-type-specific differences in chromatin architecture in complex tissues.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
MeCP2 nuclear dynamics in live neurons results from low and high affinity chromatin interactions.Elife. 2019 Dec 23;8:e51449. doi: 10.7554/eLife.51449. Elife. 2019. PMID: 31868585 Free PMC article.
-
ATRX Contributes to MeCP2-Mediated Pericentric Heterochromatin Organization during Neural Differentiation.Int J Mol Sci. 2019 Oct 29;20(21):5371. doi: 10.3390/ijms20215371. Int J Mol Sci. 2019. PMID: 31671722 Free PMC article.
-
MeCP2 Rett mutations affect large scale chromatin organization.Hum Mol Genet. 2011 Nov 1;20(21):4187-95. doi: 10.1093/hmg/ddr346. Epub 2011 Aug 10. Hum Mol Genet. 2011. PMID: 21831886
-
MeCP2 and Chromatin Compartmentalization.Cells. 2020 Apr 3;9(4):878. doi: 10.3390/cells9040878. Cells. 2020. PMID: 32260176 Free PMC article. Review.
-
The neurobiology of Rett syndrome.Neuroscientist. 2003 Feb;9(1):57-63. doi: 10.1177/1073858402239591. Neuroscientist. 2003. PMID: 12580340 Review.
Cited by
-
The anaphase-promoting complex controls a ubiquitination-phosphoprotein axis in chromatin during neurodevelopment.Dev Cell. 2023 Dec 4;58(23):2666-2683.e9. doi: 10.1016/j.devcel.2023.10.002. Epub 2023 Oct 23. Dev Cell. 2023. PMID: 37875116 Free PMC article.
-
Phase-separated chromatin compartments: Orchestrating gene expression through condensation.Cell Insight. 2024 Oct 12;3(6):100213. doi: 10.1016/j.cellin.2024.100213. eCollection 2024 Dec. Cell Insight. 2024. PMID: 39512706 Free PMC article. Review.
-
Cell-type-specific 3D-genome organization and transcription regulation in the brain.bioRxiv [Preprint]. 2023 Dec 5:2023.12.04.570024. doi: 10.1101/2023.12.04.570024. bioRxiv. 2023. PMID: 38105994 Free PMC article. Preprint.
-
MeCP2 nuclear dynamics in live neurons results from low and high affinity chromatin interactions.Elife. 2019 Dec 23;8:e51449. doi: 10.7554/eLife.51449. Elife. 2019. PMID: 31868585 Free PMC article.
-
Visualization of the spatial arrangement of nuclear organization using three-dimensional fluorescence in situ hybridization in early mouse embryos: A new "EASI-FISH chamber glass" for mammalian embryos.J Reprod Dev. 2017 Apr 21;63(2):167-174. doi: 10.1262/jrd.2016-172. Epub 2017 Feb 11. J Reprod Dev. 2017. PMID: 28190810 Free PMC article.
References
-
- Adler DA, Quaderi NA, Brown SD, Chapman VM, Moore J, Tate P, Disteche CM. The X-linked methylated DNA binding protein, Mecp2, is subject to X inactivation in the mouse. Mamm. Genome. 1995;6:491–492. - PubMed
-
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

