An Application of Sequential Meta-Analysis to Gene Expression Studies
- PMID: 26401096
- PMCID: PMC4567049
- DOI: 10.4137/CIN.S27718
An Application of Sequential Meta-Analysis to Gene Expression Studies
Abstract
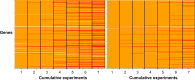
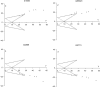
Most of the discoveries from gene expression data are driven by a study claiming an optimal subset of genes that play a key role in a specific disease. Meta-analysis of the available datasets can help in getting concordant results so that a real-life application may be more successful. Sequential meta-analysis (SMA) is an approach for combining studies in chronological order while preserving the type I error and pre-specifying the statistical power to detect a given effect size. We focus on the application of SMA to find gene expression signatures across experiments in acute myeloid leukemia. SMA of seven raw datasets is used to evaluate whether the accumulated samples show enough evidence or more experiments should be initiated. We found 313 differentially expressed genes, based on the cumulative information of the experiments. SMA offers an alternative to existing methods in generating a gene list by evaluating the adequacy of the cumulative information.
Keywords: differentially expressed genes; gene expression; sequential meta-analysis; triangular test.
Figures





Similar articles
-
Network-Based Meta-Analyses of Associations of Multiple Gene Expression Profiles with Bone Mineral Density Variations in Women.PLoS One. 2016 Jan 25;11(1):e0147475. doi: 10.1371/journal.pone.0147475. eCollection 2016. PLoS One. 2016. PMID: 26808152 Free PMC article.
-
A novel bi-level meta-analysis approach: applied to biological pathway analysis.Bioinformatics. 2016 Feb 1;32(3):409-16. doi: 10.1093/bioinformatics/btv588. Epub 2015 Oct 14. Bioinformatics. 2016. PMID: 26471455 Free PMC article.
-
Meta-analysis of microarray data: The case of imatinib resistance in chronic myelogenous leukemia.Comput Biol Chem. 2010 Jun;34(3):184-92. doi: 10.1016/j.compbiolchem.2010.06.003. Epub 2010 Jun 18. Comput Biol Chem. 2010. PMID: 20619742
-
[Transcriptomes for serial analysis of gene expression].J Soc Biol. 2002;196(4):303-7. J Soc Biol. 2002. PMID: 12645300 Review. French.
-
Acceptance and commitment therapy - Do we know enough? Cumulative and sequential meta-analyses of randomized controlled trials.J Affect Disord. 2016 Jan 15;190:551-565. doi: 10.1016/j.jad.2015.10.053. Epub 2015 Oct 30. J Affect Disord. 2016. PMID: 26571105 Review.
Cited by
-
The Role of Systems Biology in Deciphering Asthma Heterogeneity.Life (Basel). 2022 Oct 8;12(10):1562. doi: 10.3390/life12101562. Life (Basel). 2022. PMID: 36294997 Free PMC article. Review.
-
Meta-analysis approach as a gene selection method in class prediction: does it improve model performance? A case study in acute myeloid leukemia.BMC Bioinformatics. 2017 Apr 11;18(1):210. doi: 10.1186/s12859-017-1619-7. BMC Bioinformatics. 2017. PMID: 28399794 Free PMC article.
-
Derangement of cell cycle markers in peripheral blood mononuclear cells of asthmatic patients as a reliable biomarker for asthma control.Sci Rep. 2021 Jun 4;11(1):11873. doi: 10.1038/s41598-021-91087-5. Sci Rep. 2021. PMID: 34088958 Free PMC article.
References
-
- Catherino WH, Segars JH. Microarray analysis in fibroids: which gene list is the correct list? Fertil Steril. 2003;80(2):293–4. - PubMed
-
- Fortunel NO, Otu HH, Ng HH, et al. Comment on “‘Stemness’: transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature”. Science. 2003;302(5644):393. author reply 393. - PubMed
-
- Evsikov AV, Solter D. Comment on “‘Stemness’: transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature”. Science. 2003;302(5644):393. author reply 393. - PubMed
LinkOut - more resources
Full Text Sources

