NSun2 Promotes Cell Growth via Elevating Cyclin-Dependent Kinase 1 Translation
- PMID: 26391950
- PMCID: PMC4628067
- DOI: 10.1128/MCB.00742-15
NSun2 Promotes Cell Growth via Elevating Cyclin-Dependent Kinase 1 Translation
Abstract
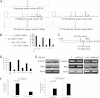
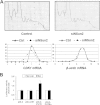
The tRNA methytransferase NSun2 promotes cell proliferation, but the molecular mechanism has not been elucidated. Here, we report that NSun2 regulates cyclin-dependent kinase 1 (CDK1) expression in a cell cycle-dependent manner. Knockdown of NSun2 decreased the CDK1 protein level, while overexpression of NSun2 elevated it without altering CDK1 mRNA levels. Further studies revealed that NSun2 methylated CDK1 mRNA in vitro and in cells and that methylation by NSun2 enhanced CDK1 translation. Importantly, NSun2-mediated regulation of CDK1 expression had an impact on the cell division cycle. These results provide new insight into the regulation of CDK1 during the cell division cycle.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation.Aging (Albany NY). 2015 Dec;7(12):1143-58. doi: 10.18632/aging.100860. Aging (Albany NY). 2015. PMID: 26687548 Free PMC article.
-
RNA methyltransferase NSUN2 promotes stress-induced HUVEC senescence.Oncotarget. 2016 Apr 12;7(15):19099-110. doi: 10.18632/oncotarget.8087. Oncotarget. 2016. PMID: 26992231 Free PMC article.
-
The tRNA methyltransferase NSun2 stabilizes p16INK⁴ mRNA by methylating the 3'-untranslated region of p16.Nat Commun. 2012 Mar 6;3:712. doi: 10.1038/ncomms1692. Nat Commun. 2012. PMID: 22395603 Free PMC article.
-
mRNA methylation by NSUN2 in cell proliferation.Wiley Interdiscip Rev RNA. 2016 Nov;7(6):838-842. doi: 10.1002/wrna.1380. Epub 2016 Jul 22. Wiley Interdiscip Rev RNA. 2016. PMID: 27444144 Review.
-
Translational control during mitosis.Biochimie. 2005 Sep-Oct;87(9-10):805-11. doi: 10.1016/j.biochi.2005.04.014. Biochimie. 2005. PMID: 15951098 Review.
Cited by
-
The RNA Methyltransferase NSUN2 and Its Potential Roles in Cancer.Cells. 2020 Jul 22;9(8):1758. doi: 10.3390/cells9081758. Cells. 2020. PMID: 32708015 Free PMC article. Review.
-
Roles of RNA Modifications in Diverse Cellular Functions.Front Cell Dev Biol. 2022 Mar 8;10:828683. doi: 10.3389/fcell.2022.828683. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35350378 Free PMC article. Review.
-
Chemical modifications to mRNA nucleobases impact translation elongation and termination.Biophys Chem. 2022 Jun;285:106780. doi: 10.1016/j.bpc.2022.106780. Epub 2022 Feb 16. Biophys Chem. 2022. PMID: 35313212 Free PMC article. Review.
-
Expression profiles of long noncoding RNAs associated with the NSUN2 gene in HepG2 cells.Mol Med Rep. 2019 Apr;19(4):2999-3008. doi: 10.3892/mmr.2019.9984. Epub 2019 Feb 25. Mol Med Rep. 2019. PMID: 30816500 Free PMC article.
-
Aberrant NSUN2-mediated m5C modification of exosomal LncRNA MALAT1 induced RANKL-mediated bone destruction in multiple myeloma.Commun Biol. 2024 Oct 2;7(1):1249. doi: 10.1038/s42003-024-06918-8. Commun Biol. 2024. PMID: 39358426 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
