ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13
- PMID: 26370498
- PMCID: PMC4576869
- DOI: 10.1083/jcb.201502105
ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13
Abstract
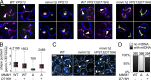
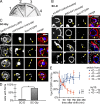
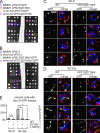
The endoplasmic reticulum-mitochondria encounter structure (ERMES) complex tethers the endoplasmic reticulum and the mitochondria. It is thought to facilitate interorganelle lipid exchange and influence mitochondrial dynamics and mitochondrial DNA maintenance. Despite this important role, ERMES is not found in metazoans. Here, we identified single amino acid substitutions in Vps13 (vacuolar protein sorting 13), a large universally conserved eukaryotic protein, which suppress all measured phenotypic consequences of ERMES deficiency. Combined loss of VPS13 and ERMES is lethal, indicating that Vps13 and ERMES function in redundant pathways. Vps13 dynamically localizes to vacuole-mitochondria and to vacuole-nucleus contact sites depending on growth conditions, suggesting that ERMES function can be bypassed by the activity of other contact sites, and that contact sites establish a growth condition-regulated organelle network.
© 2015 Lang et al.
Figures

Similar articles
-
Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites.Mol Biol Cell. 2016 Aug 1;27(15):2435-49. doi: 10.1091/mbc.E16-02-0112. Epub 2016 Jun 8. Mol Biol Cell. 2016. PMID: 27280386 Free PMC article.
-
Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites.J Cell Biol. 2017 Oct 2;216(10):3219-3229. doi: 10.1083/jcb.201610055. Epub 2017 Sep 1. J Cell Biol. 2017. PMID: 28864540 Free PMC article.
-
Endoplasmic reticulum-mitochondria junction is required for iron homeostasis.J Biol Chem. 2017 Aug 11;292(32):13197-13204. doi: 10.1074/jbc.M117.784249. Epub 2017 Jun 21. J Biol Chem. 2017. PMID: 28637866 Free PMC article.
-
The ERMES complex and ER-mitochondria connections.Biochem Soc Trans. 2012 Apr;40(2):445-50. doi: 10.1042/BST20110758. Biochem Soc Trans. 2012. PMID: 22435828 Review.
-
Organelle contact zones as sites for lipid transfer.J Biochem. 2019 Feb 1;165(2):115-123. doi: 10.1093/jb/mvy088. J Biochem. 2019. PMID: 30371789 Review.
Cited by
-
Systematic analysis of nuclear gene function in respiratory growth and expression of the mitochondrial genome in S. cerevisiae.Microb Cell. 2020 Jun 30;7(9):234-249. doi: 10.15698/mic2020.09.729. Microb Cell. 2020. PMID: 32904421 Free PMC article.
-
Meiotic Cytokinesis in Saccharomyces cerevisiae: Spores That Just Need Closure.J Fungi (Basel). 2024 Feb 6;10(2):132. doi: 10.3390/jof10020132. J Fungi (Basel). 2024. PMID: 38392804 Free PMC article. Review.
-
Remodelling of Nucleus-Vacuole Junctions During Metabolic and Proteostatic Stress.Contact (Thousand Oaks). 2021 May 27;4:25152564211016608. doi: 10.1177/25152564211016608. eCollection 2021 Jan-Dec. Contact (Thousand Oaks). 2021. PMID: 34124572 Free PMC article.
-
VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites.J Cell Biol. 2018 Oct 1;217(10):3625-3639. doi: 10.1083/jcb.201807019. Epub 2018 Aug 9. J Cell Biol. 2018. PMID: 30093493 Free PMC article.
-
VPS13: A lipid transfer protein making contacts at multiple cellular locations.J Cell Biol. 2018 Oct 1;217(10):3322-3324. doi: 10.1083/jcb.201808151. Epub 2018 Sep 4. J Cell Biol. 2018. PMID: 30181317 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

