Proteomic analysis of HIV-1 Gag interacting partners using proximity-dependent biotinylation
- PMID: 26362536
- PMCID: PMC4566291
- DOI: 10.1186/s12985-015-0365-6
Proteomic analysis of HIV-1 Gag interacting partners using proximity-dependent biotinylation
Abstract
Background: The human immunodeficiency virus type 1 (HIV-1) Gag polyprotein is necessary and sufficient to assemble non-infectious particles. Given that HIV-1 subverts many host proteins at all stages of its life cycle, it is essential to identify these interactions as potential targets for antiretroviral therapy.
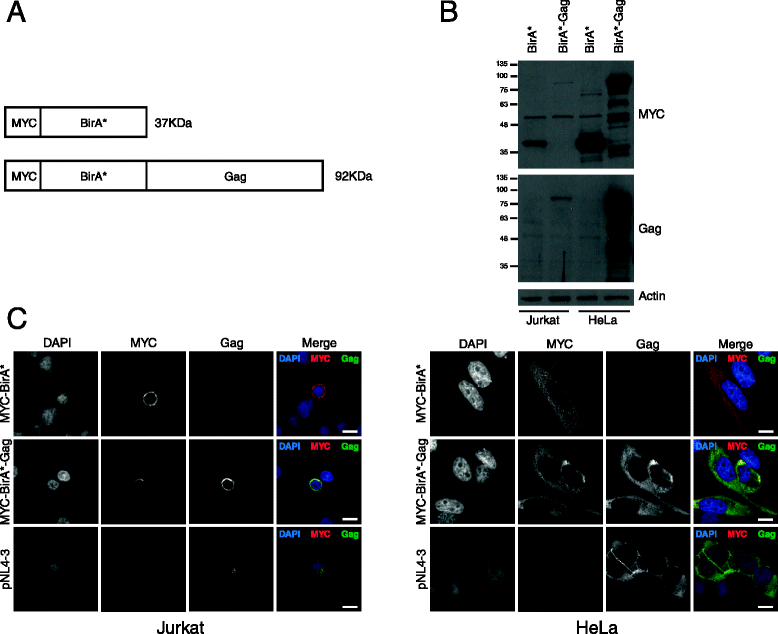
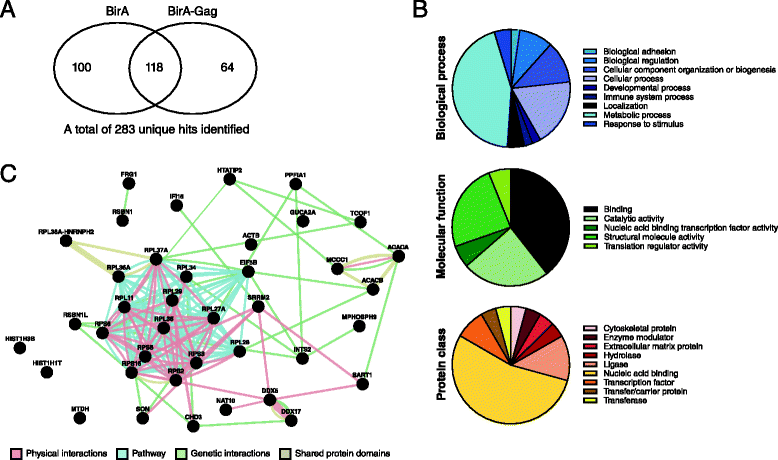
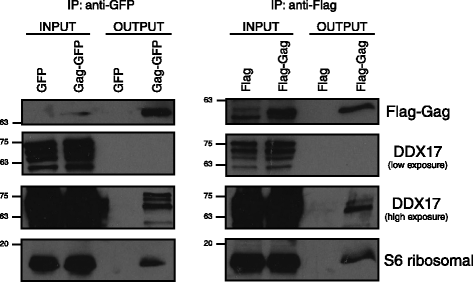
Findings: This work demonstrates the use of proximity-dependent biotin identification (BioID) of host proteins and complexes that are proximal to the N-terminal domains of the HIV-1 Gag polyprotein. Two of the hits identified in the BioID screen were validated by immunoprecipation and confirmed the interaction of DDX17 and RPS6 with HIV-1 Gag.
Conclusions: Our results show that BioID is both a successful and complementary method to screen for nearby interacting proteins of HIV-1 Gag during the replicative cycle in different cell lines.
Figures
Similar articles
-
Tandem immunoprecipitation approach to identify HIV-1 Gag associated host factors.J Virol Methods. 2014 Jul;203:116-9. doi: 10.1016/j.jviromet.2014.03.017. Epub 2014 Mar 30. J Virol Methods. 2014. PMID: 24690621
-
Analysis of HIV-1 Gag protein interactions via biotin ligase tagging.J Virol. 2015 Apr;89(7):3988-4001. doi: 10.1128/JVI.03584-14. Epub 2015 Jan 28. J Virol. 2015. PMID: 25631074 Free PMC article.
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y. Retrovirology. 2024. PMID: 38898526 Free PMC article.
-
Roles of the interactions between Env and Gag proteins in the HIV-1 replication cycle.Microbiol Immunol. 2008 May;52(5):287-95. doi: 10.1111/j.1348-0421.2008.00008.x. Microbiol Immunol. 2008. PMID: 18557900 Review.
-
HIV-1 gag proteins: diverse functions in the virus life cycle.Virology. 1998 Nov 10;251(1):1-15. doi: 10.1006/viro.1998.9398. Virology. 1998. PMID: 9813197 Review.
Cited by
-
Target Discovery for Host-Directed Antiviral Therapies: Application of Proteomics Approaches.mSystems. 2021 Oct 26;6(5):e0038821. doi: 10.1128/mSystems.00388-21. Epub 2021 Sep 14. mSystems. 2021. PMID: 34519533 Free PMC article.
-
DDX17 Specifically, and Independently of DDX5, Controls Use of the HIV A4/5 Splice Acceptor Cluster and Is Essential for Efficient Replication of HIV.J Mol Biol. 2018 Sep 14;430(18 Pt B):3111-3128. doi: 10.1016/j.jmb.2018.06.052. Epub 2018 Jul 2. J Mol Biol. 2018. PMID: 30131116 Free PMC article.
-
Microbial Pathogenesis in the Era of Spatial Omics.Infect Immun. 2023 Jul 18;91(7):e0044222. doi: 10.1128/iai.00442-22. Epub 2023 May 31. Infect Immun. 2023. PMID: 37255461 Free PMC article. Review.
-
Quantitative Interactomics of Lck-TurboID in Living Human T Cells Unveils T Cell Receptor Stimulation-Induced Proximal Lck Interactors.J Proteome Res. 2021 Jan 1;20(1):715-726. doi: 10.1021/acs.jproteome.0c00616. Epub 2020 Nov 13. J Proteome Res. 2021. PMID: 33185455 Free PMC article.
-
Leveraging biotin-based proximity labeling to identify cellular factors governing early alphaherpesvirus infection.mBio. 2024 Aug 14;15(8):e0144524. doi: 10.1128/mbio.01445-24. Epub 2024 Jul 2. mBio. 2024. PMID: 38953638 Free PMC article.
References
-
- Petry H, Goldmann C, Ast O, Luke W. The use of virus-like particles for gene transfer. Curr Opin Mol Ther. 2003;5(5):524–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

