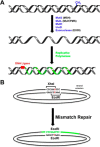
Mismatch repair
- PMID: 26354434
- PMCID: PMC4646297
- DOI: 10.1074/jbc.R115.660142
Mismatch repair
Abstract
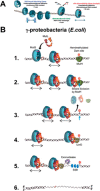
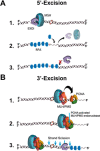
Highly conserved MutS homologs (MSH) and MutL homologs (MLH/PMS) are the fundamental components of mismatch repair (MMR). After decades of debate, it appears clear that the MSH proteins initiate MMR by recognizing a mismatch and forming multiple extremely stable ATP-bound sliding clamps that diffuse without hydrolysis along the adjacent DNA. The function(s) of MLH/PMS proteins is less clear, although they too bind ATP and are targeted to MMR by MSH sliding clamps. Structural analysis combined with recent real-time single molecule and cellular imaging technologies are providing new and detailed insight into the thermal-driven motions that animate the complete MMR mechanism.
Keywords: DNA mismatch repair; DNA repair; Lynch syndrome; cancer biology; hMLH1; hMSH2; hereditary non-polyposis colorectal cancer; mutagenesis; single molecule analysis; single-molecule biophysics.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Similar articles
-
Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair.Nature. 2016 Nov 24;539(7630):583-587. doi: 10.1038/nature20562. Epub 2016 Nov 16. Nature. 2016. PMID: 27851738 Free PMC article.
-
MutS homolog sliding clamps shield the DNA from binding proteins.J Biol Chem. 2018 Sep 14;293(37):14285-14294. doi: 10.1074/jbc.RA118.002264. Epub 2018 Aug 2. J Biol Chem. 2018. PMID: 30072380 Free PMC article.
-
Dynamic control of strand excision during human DNA mismatch repair.Proc Natl Acad Sci U S A. 2016 Mar 22;113(12):3281-6. doi: 10.1073/pnas.1523748113. Epub 2016 Mar 7. Proc Natl Acad Sci U S A. 2016. PMID: 26951673 Free PMC article.
-
Single-molecule views of MutS on mismatched DNA.DNA Repair (Amst). 2014 Aug;20:82-93. doi: 10.1016/j.dnarep.2014.02.014. Epub 2014 Mar 12. DNA Repair (Amst). 2014. PMID: 24629484 Free PMC article. Review.
-
Stochastic Processes and Component Plasticity Governing DNA Mismatch Repair.J Mol Biol. 2018 Oct 26;430(22):4456-4468. doi: 10.1016/j.jmb.2018.05.039. Epub 2018 Jun 1. J Mol Biol. 2018. PMID: 29864444 Free PMC article. Review.
Cited by
-
Targeting the DNA damage response in cancer.MedComm (2020). 2024 Oct 31;5(11):e788. doi: 10.1002/mco2.788. eCollection 2024 Nov. MedComm (2020). 2024. PMID: 39492835 Free PMC article. Review.
-
HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins.Proc Natl Acad Sci U S A. 2016 Jul 5;113(27):E3921-30. doi: 10.1073/pnas.1605023113. Epub 2016 Jun 22. Proc Natl Acad Sci U S A. 2016. PMID: 27335459 Free PMC article.
-
Dianhydrogalactitol synergizes with topoisomerase poisons to overcome DNA repair activity in tumor cells.Cell Death Dis. 2020 Jul 24;11(7):577. doi: 10.1038/s41419-020-02780-8. Cell Death Dis. 2020. PMID: 32709853 Free PMC article.
-
The ubiquitin ligase UBR4 and the deubiquitylase USP5 modulate the stability of DNA mismatch repair protein MLH1.J Biol Chem. 2024 Aug;300(8):107592. doi: 10.1016/j.jbc.2024.107592. Epub 2024 Jul 18. J Biol Chem. 2024. PMID: 39032648 Free PMC article.
-
Mutation of TGFβ-RII eliminates NSAID cancer chemoprevention.Oncotarget. 2017 Dec 31;9(16):12554-12561. doi: 10.18632/oncotarget.23792. eCollection 2018 Feb 27. Oncotarget. 2017. PMID: 29560090 Free PMC article.
References
-
- Witkin E. M., and Sicurella N. A. (1964) Pure clones of lactose negative mutants obtained in Escherichia coli after treatment with 5-bromouracil. J. Mol. Biol. 8, 610–613 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

