Phage display technique identifies the interaction of severe acute respiratory syndrome coronavirus open reading frame 6 protein with nuclear pore complex interacting protein NPIPB3 in modulating Type I interferon antagonism
- PMID: 26320399
- PMCID: PMC7105049
- DOI: 10.1016/j.jmii.2015.07.002
Phage display technique identifies the interaction of severe acute respiratory syndrome coronavirus open reading frame 6 protein with nuclear pore complex interacting protein NPIPB3 in modulating Type I interferon antagonism
Abstract
Background/purpose: Severe acute respiratory syndrome coronavirus (SARS-CoV) proteins including ORF6 inhibit Type I interferon (IFN) signaling.
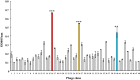
Methods: This study identified SARS-CoV ORF6-interacting proteins using the phage displayed human lung cDNA libraries, and examined the association of ORF6-host factor interaction with Type I IFN antagonism. After the fifth round of biopanning with Escherichia coli-synthesized ORF6-His tagged protein, the relative binding affinity of phage clones to ORF6 was determined using direct enzyme-linked immunosorbent assay.
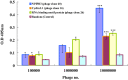
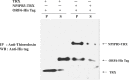
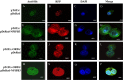
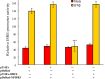
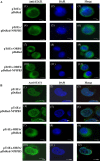
Results: The highest affinity clone to ORF6 displayed the C-terminal domain of NPIPB3 (nuclear pore complex interacting protein family, member B3; also named as phosphatidylinositol-3-kinase-related kinase SMG-1 isoform 1 homolog). The coimmunoprecipitation assay demonstrated the direct binding of ORF6 to the C-terminal domain of NPIPB3 in vitro. Confocal imaging revealed a close colocalization of SARS-CoV ORF6 protein with NPIPB3 in human promonocytes. The dual luciferase reporter assay showed that the C-terminal domain of NPIPB3 attenuated the antagonistic activity of SARS-CoV ORF6 on IFN-β-induced ISRE (IFN stimulated response element)-responsive firefly luciferase activity. In addition, confocal imaging and Western blotting assays revealed that the increases in STAT-1 nuclear translocation and phosphorylation occurred in the transfected cells expressing both genes of ORF6 and NPIPB3, but not in the ORF6-expressing cells in response to IFN-β.
Conclusion: The overexpression of NPIPB3 restored the IFN-β responses in SARS-CoV ORF6 expressing cells, indicating that the interaction of SARS CoV ORF6 and NPIPB3 reduced Type I IFN antagonism by SARS-CoV ORF6.
Keywords: IFN antagonism; NPIPB3; ORF6; SARS-CoV; phage display.
Copyright © 2015. Published by Elsevier B.V.
Figures


Similar articles
-
Interferon antagonism by SARS-CoV-2: a functional study using reverse genetics.Lancet Microbe. 2021 May;2(5):e210-e218. doi: 10.1016/S2666-5247(21)00027-6. Epub 2021 Mar 4. Lancet Microbe. 2021. PMID: 33969329 Free PMC article.
-
SARS-CoV-2 ORF6 protein does not antagonize interferon signaling in respiratory epithelial Calu-3 cells during infection.mBio. 2023 Aug 31;14(4):e0119423. doi: 10.1128/mbio.01194-23. Epub 2023 Jun 28. mBio. 2023. PMID: 37377442 Free PMC article.
-
SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling.Proc Natl Acad Sci U S A. 2020 Nov 10;117(45):28344-28354. doi: 10.1073/pnas.2016650117. Epub 2020 Oct 23. Proc Natl Acad Sci U S A. 2020. PMID: 33097660 Free PMC article.
-
Roles and functions of SARS-CoV-2 proteins in host immune evasion.Front Immunol. 2022 Aug 8;13:940756. doi: 10.3389/fimmu.2022.940756. eCollection 2022. Front Immunol. 2022. PMID: 36003396 Free PMC article. Review.
-
Spatial and temporal roles of SARS-CoV PLpro -A snapshot.FASEB J. 2021 Jan;35(1):e21197. doi: 10.1096/fj.202002271. FASEB J. 2021. PMID: 33368679 Free PMC article. Review.
Cited by
-
SARS-CoV-2 Nsp8 suppresses MDA5 antiviral immune responses by impairing TRIM4-mediated K63-linked polyubiquitination.PLoS Pathog. 2023 Nov 13;19(11):e1011792. doi: 10.1371/journal.ppat.1011792. eCollection 2023 Nov. PLoS Pathog. 2023. PMID: 37956198 Free PMC article.
-
Genomic regions associated with microdeletion/microduplication syndromes exhibit extreme diversity of structural variation.Genetics. 2021 Feb 9;217(2):iyaa038. doi: 10.1093/genetics/iyaa038. Genetics. 2021. PMID: 33724415 Free PMC article.
-
Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures.J Virol. 2020 Sep 15;94(19):e00985-20. doi: 10.1128/JVI.00985-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32699094 Free PMC article.
-
Phage Display Technique as a Tool for Diagnosis and Antibody Selection for Coronaviruses.Curr Microbiol. 2021 Apr;78(4):1124-1134. doi: 10.1007/s00284-021-02398-9. Epub 2021 Mar 9. Curr Microbiol. 2021. PMID: 33687511 Free PMC article. Review.
-
Human core duplicon gene families: game changers or game players?Brief Funct Genomics. 2019 Nov 19;18(6):402-411. doi: 10.1093/bfgp/elz016. Brief Funct Genomics. 2019. PMID: 31529038 Free PMC article.
References
-
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. - PubMed
-
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

