DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts
- PMID: 26317465
- PMCID: PMC4843502
- DOI: 10.1016/j.cell.2015.07.056
DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts
Abstract
DNA-demethylating agents have shown clinical anti-tumor efficacy via an unknown mechanism of action. Using a combination of experimental and bioinformatics analyses in colorectal cancer cells, we demonstrate that low-dose 5-AZA-CdR targets colorectal cancer-initiating cells (CICs) by inducing viral mimicry. This is associated with induction of dsRNAs derived at least in part from endogenous retroviral elements, activation of the MDA5/MAVS RNA recognition pathway, and downstream activation of IRF7. Indeed, disruption of virus recognition pathways, by individually knocking down MDA5, MAVS, or IRF7, inhibits the ability of 5-AZA-CdR to target colorectal CICs and significantly decreases 5-AZA-CdR long-term growth effects. Moreover, transfection of dsRNA into CICs can mimic the effects of 5-AZA-CdR. Together, our results represent a major shift in understanding the anti-tumor mechanisms of DNA-demethylating agents and highlight the MDA5/MAVS/IRF7 pathway as a potentially druggable target against CICs.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


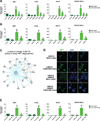
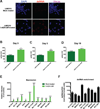
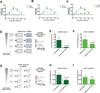
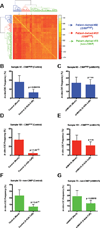

Comment in
-
DNA Methylation Inhibitors in Cancer Therapy: The Immunity Dimension.Cell. 2015 Aug 27;162(5):938-9. doi: 10.1016/j.cell.2015.08.005. Cell. 2015. PMID: 26317460
Similar articles
-
MAVS-dependent IRF3/7 bypass of interferon β-induction restricts the response to measles infection in CD150Tg mouse bone marrow-derived dendritic cells.Mol Immunol. 2014 Feb;57(2):100-10. doi: 10.1016/j.molimm.2013.08.007. Epub 2013 Oct 4. Mol Immunol. 2014. PMID: 24096085
-
DNAJB1/HSP40 Suppresses Melanoma Differentiation-Associated Gene 5-Mitochondrial Antiviral Signaling Protein Function in Conjunction with HSP70.J Innate Immun. 2018;10(1):44-55. doi: 10.1159/000480740. Epub 2017 Oct 26. J Innate Immun. 2018. PMID: 29069650 Free PMC article.
-
DNA Methylation Inhibitors in Cancer Therapy: The Immunity Dimension.Cell. 2015 Aug 27;162(5):938-9. doi: 10.1016/j.cell.2015.08.005. Cell. 2015. PMID: 26317460
-
Pharmacological approach for optimization of the dose schedule of 5-Aza-2'-deoxycytidine (Decitabine) for the therapy of leukemia.Leukemia. 1997 Feb;11(2):175-80. doi: 10.1038/sj.leu.2400550. Leukemia. 1997. PMID: 9009076 Review.
-
Pharmacological approach for optimization of the dose schedule of 5-Aza-2'-deoxycytidine (Decitabine) for the therapy of leukemia.Leukemia. 1997 Mar;11 Suppl 1:S1-6. Leukemia. 1997. PMID: 9130684 Review.
Cited by
-
In vivo CRISPR screens identify a dual function of MEN1 in regulating tumor-microenvironment interactions.Nat Genet. 2024 Sep;56(9):1890-1902. doi: 10.1038/s41588-024-01874-9. Epub 2024 Sep 3. Nat Genet. 2024. PMID: 39227744 Free PMC article.
-
microRNA-mediated resistance to hypoglycemia in the HepG2 human hepatoma cell line.BMC Cancer. 2016 Sep 15;16(1):732. doi: 10.1186/s12885-016-2762-7. BMC Cancer. 2016. PMID: 27629773 Free PMC article.
-
Reciprocal regulation of RIG-I and XRCC4 connects DNA repair with RIG-I immune signaling.Nat Commun. 2021 Apr 12;12(1):2187. doi: 10.1038/s41467-021-22484-7. Nat Commun. 2021. PMID: 33846346 Free PMC article.
-
Targeting epigenetic and posttranslational modifications regulating ferroptosis for the treatment of diseases.Signal Transduct Target Ther. 2023 Dec 10;8(1):449. doi: 10.1038/s41392-023-01720-0. Signal Transduct Target Ther. 2023. PMID: 38072908 Free PMC article. Review.
-
Ancient Adversary - HERV-K (HML-2) in Cancer.Front Oncol. 2021 May 13;11:658489. doi: 10.3389/fonc.2021.658489. eCollection 2021. Front Oncol. 2021. PMID: 34055625 Free PMC article. Review.
References
-
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annual review of immunology. 2011;29:185–214. - PubMed
-
- Besch R, Poeck H, Hohenauer T, Senft D, Hacker G, Berking C, Hornung V, Endres S, Ruzicka T, Rothenfusser S, et al. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. The Journal of clinical investigation. 2009;119:2399–2411. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

