MacroH2A1 and ATM Play Opposing Roles in Paracrine Senescence and the Senescence-Associated Secretory Phenotype
- PMID: 26300260
- PMCID: PMC4548812
- DOI: 10.1016/j.molcel.2015.07.011
MacroH2A1 and ATM Play Opposing Roles in Paracrine Senescence and the Senescence-Associated Secretory Phenotype
Abstract
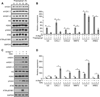
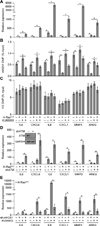
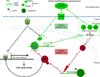
Oncogene-induced senescence (OIS) is a tumor-suppressive mechanism typified by stable proliferative arrest, a persistent DNA damage response, and the senescence-associated secretory phenotype (SASP), which helps to maintain the senescent state and triggers bystander senescence in a paracrine fashion. Here, we demonstrate that the tumor suppressive histone variant macroH2A1 is a critical component of the positive feedback loop that maintains SASP gene expression and triggers the induction of paracrine senescence. MacroH2A1 undergoes dramatic genome-wide relocalization during OIS, including its removal from SASP gene chromatin. The removal of macroH2A1 from SASP genes results from a negative feedback loop activated by SASP-mediated endoplasmic reticulum (ER) stress. ER stress leads to increased reactive oxygen species and persistent DNA damage response including activation of ATM, which mediates removal macroH2A1 from SASP genes. Together, our findings indicate that macroH2A1 is a critical control point for the regulation of SASP gene expression during senescence.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
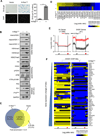
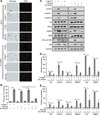
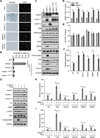
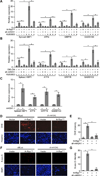
Comment in
-
ATM, MacroH2A.1, and SASP: The Checks and Balances of Cellular Senescence.Mol Cell. 2015 Sep 3;59(5):713-5. doi: 10.1016/j.molcel.2015.08.010. Mol Cell. 2015. PMID: 26340421
Similar articles
-
DNA Hypomethylation and Histone Variant macroH2A1 Synergistically Attenuate Chemotherapy-Induced Senescence to Promote Hepatocellular Carcinoma Progression.Cancer Res. 2016 Feb 1;76(3):594-606. doi: 10.1158/0008-5472.CAN-15-1336. Epub 2016 Jan 15. Cancer Res. 2016. PMID: 26772755 Free PMC article.
-
IκBζ is a regulator of the senescence-associated secretory phenotype in DNA damage- and oncogene-induced senescence.J Cell Sci. 2013 Aug 15;126(Pt 16):3738-45. doi: 10.1242/jcs.128835. Epub 2013 Jun 18. J Cell Sci. 2013. PMID: 23781024
-
MLL1 is essential for the senescence-associated secretory phenotype.Genes Dev. 2016 Feb 1;30(3):321-36. doi: 10.1101/gad.271882.115. Genes Dev. 2016. PMID: 26833731 Free PMC article.
-
Emerging role of NF-κB signaling in the induction of senescence-associated secretory phenotype (SASP).Cell Signal. 2012 Apr;24(4):835-45. doi: 10.1016/j.cellsig.2011.12.006. Epub 2011 Dec 11. Cell Signal. 2012. PMID: 22182507 Review.
-
The Senescence-Associated Secretory Phenotype: Critical Effector in Skin Cancer and Aging.J Invest Dermatol. 2016 Nov;136(11):2133-2139. doi: 10.1016/j.jid.2016.06.621. Epub 2016 Aug 17. J Invest Dermatol. 2016. PMID: 27543988 Free PMC article. Review.
Cited by
-
The histone variant macroH2A1 is a splicing-modulated caretaker of genome integrity and tumor growth.Mol Cell Oncol. 2018 Apr 4;5(3):e1441629. doi: 10.1080/23723556.2018.1441629. eCollection 2018. Mol Cell Oncol. 2018. PMID: 30250894 Free PMC article.
-
Macro Histone Variants: Emerging Rheostats of Gastrointestinal Cancers.Cancers (Basel). 2019 May 15;11(5):676. doi: 10.3390/cancers11050676. Cancers (Basel). 2019. PMID: 31096699 Free PMC article. Review.
-
Mitochondrial Homeostasis and Cellular Senescence.Cells. 2019 Jul 6;8(7):686. doi: 10.3390/cells8070686. Cells. 2019. PMID: 31284597 Free PMC article. Review.
-
In medio stat virtus: unanticipated consequences of telomere dysequilibrium.Philos Trans R Soc Lond B Biol Sci. 2018 Mar 5;373(1741):20160444. doi: 10.1098/rstb.2016.0444. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 29335368 Free PMC article. Review.
-
Exploring Transcriptional Regulation of Beta Cell SASP by Brd4-Associated Proteins and Cell Cycle Control Protein p21.Epigenomes. 2024 Mar 6;8(1):10. doi: 10.3390/epigenomes8010010. Epigenomes. 2024. PMID: 38534794 Free PMC article.
References
-
- Acosta J, O’Loghlen A, Banito A, Raguz S, Gil J. Control of senescence by CXCR2 and its ligands. Cell Cycle. 2008a;7:2956–2959. - PubMed
-
- Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008b;133:1006–1018. - PubMed
-
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

