Knock-In Mice with NOP-eGFP Receptors Identify Receptor Cellular and Regional Localization
- PMID: 26290245
- PMCID: PMC4540802
- DOI: 10.1523/JNEUROSCI.5122-14.2015
Knock-In Mice with NOP-eGFP Receptors Identify Receptor Cellular and Regional Localization
Abstract
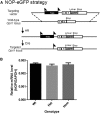
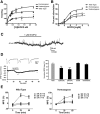
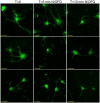
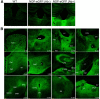
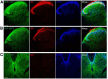
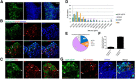
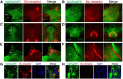
The nociceptin/orphanin FQ (NOP) receptor, the fourth member of the opioid receptor family, is involved in many processes common to the opioid receptors including pain and drug abuse. To better characterize receptor location and trafficking, knock-in mice were created by inserting the gene encoding enhanced green fluorescent protein (eGFP) into the NOP receptor gene (Oprl1) and producing mice expressing a functional NOP-eGFP C-terminal fusion in place of the native NOP receptor. The NOP-eGFP receptor was present in brain of homozygous knock-in animals in concentrations somewhat higher than in wild-type mice and was functional when tested for stimulation of [(35)S]GTPγS binding in vitro and in patch-clamp electrophysiology in dorsal root ganglia (DRG) neurons and hippocampal slices. Inhibition of morphine analgesia was equivalent when tested in knock-in and wild-type mice. Imaging revealed detailed neuroanatomy in brain, spinal cord, and DRG and was generally consistent with in vitro autoradiographic imaging of receptor location. Multicolor immunohistochemistry identified cells coexpressing various spinal cord and DRG cellular markers, as well as coexpression with μ-opioid receptors in DRG and brain regions. Both in tissue slices and primary cultures, the NOP-eGFP receptors appear throughout the cell body and in processes. These knock-in mice have NOP receptors that function both in vitro and in vivo and appear to be an exceptional tool to study receptor neuroanatomy and correlate with NOP receptor function.
Significance statement: The NOP receptor, the fourth member of the opioid receptor family, is involved in pain, drug abuse, and a number of other CNS processes. The regional and cellular distribution has been difficult to determine due to lack of validated antibodies for immunohistochemical analysis. To provide a new tool for the investigation of receptor localization, we have produced knock-in mice with a fluorescent-tagged NOP receptor in place of the native NOP receptor. These knock-in mice have NOP receptors that function both in vitro and in vivo and have provided a detailed characterization of NOP receptors in brain, spinal cord, and DRG neurons. They appear to be an exceptional tool to study receptor neuroanatomy and correlate with NOP receptor function.
Keywords: GPCR; N/OFQ; NOP receptor; eGFP; histochemistry; knock-in.
Copyright © 2015 the authors 0270-6474/15/3511683-12$15.00/0.
Figures
Similar articles
-
Analysis of the distribution of spinal NOP receptors in a chronic pain model using NOP-eGFP knock-in mice.Br J Pharmacol. 2018 Jul;175(13):2662-2675. doi: 10.1111/bph.14225. Epub 2018 May 6. Br J Pharmacol. 2018. PMID: 29582417 Free PMC article.
-
Nociceptin/orphanin FQ receptor expression in clinical pain disorders and functional effects in cultured neurons.Pain. 2016 Sep;157(9):1960-1969. doi: 10.1097/j.pain.0000000000000597. Pain. 2016. PMID: 27127846
-
The biology of Nociceptin/Orphanin FQ (N/OFQ) related to obesity, stress, anxiety, mood, and drug dependence.Pharmacol Ther. 2014 Mar;141(3):283-99. doi: 10.1016/j.pharmthera.2013.10.011. Epub 2013 Nov 1. Pharmacol Ther. 2014. PMID: 24189487 Free PMC article. Review.
-
Ro 64-6198 [(1S,3aS)-8-(2,3,3a,4,5,6-Hexahydro-1H-phenalen-1-yl)-1-phenyl-1,3,8-triaza-spiro[4.5]decan-4-one] acts differently from nociceptin/orphanin FQ in rat periaqueductal gray slices.J Pharmacol Exp Ther. 2004 Nov;311(2):645-51. doi: 10.1124/jpet.104.070219. Epub 2004 Jul 13. J Pharmacol Exp Ther. 2004. PMID: 15254141
-
Pharmacological characterization of nociceptin/orphanin FQ receptors, a novel opioid receptor family, in the midbrain periaqueductal gray.Ann N Y Acad Sci. 2004 Oct;1025:398-403. doi: 10.1196/annals.1316.049. Ann N Y Acad Sci. 2004. PMID: 15542742 Review.
Cited by
-
Targeting Nociceptin/Orphanin FQ receptor to rescue cognitive symptoms in a mouse neuroendocrine model of chronic stress.Mol Psychiatry. 2024 Mar;29(3):718-729. doi: 10.1038/s41380-023-02363-x. Epub 2023 Dec 20. Mol Psychiatry. 2024. PMID: 38123728
-
Analysis of the distribution of spinal NOP receptors in a chronic pain model using NOP-eGFP knock-in mice.Br J Pharmacol. 2018 Jul;175(13):2662-2675. doi: 10.1111/bph.14225. Epub 2018 May 6. Br J Pharmacol. 2018. PMID: 29582417 Free PMC article.
-
HA-MOP knockin mice express the canonical µ-opioid receptor but lack detectable splice variants.Commun Biol. 2021 Sep 14;4(1):1070. doi: 10.1038/s42003-021-02580-6. Commun Biol. 2021. PMID: 34522000 Free PMC article.
-
Expression of Opioid Receptors in Cells of the Immune System.Int J Mol Sci. 2020 Dec 30;22(1):315. doi: 10.3390/ijms22010315. Int J Mol Sci. 2020. PMID: 33396783 Free PMC article. Review.
-
Spotlight on Nociceptin/Orphanin FQ Receptor in the Treatment of Pain.Molecules. 2022 Jan 18;27(3):595. doi: 10.3390/molecules27030595. Molecules. 2022. PMID: 35163856 Free PMC article. Review.
References
-
- Bardoni R, Tawfik VL, Wang D, François A, Solorzano C, Shuster SA, Choudhury P, Betelli C, Cassidy C, Smith K, de Nooij JC, Mennicken F, O'Donnell D, Kieffer BL, Woodbury CJ, Basbaum AI, MacDermott AB, Scherrer G. Delta opioid receptors presynaptically regulate cutaneous mechanosensory neuron input to the spinal cord dorsal horn. Neuron. 2014;81:1312–1327. doi: 10.1016/j.neuron.2014.01.044. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
