Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice
- PMID: 26274050
- PMCID: PMC4737925
- DOI: 10.1038/ismej.2015.114
Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice
Abstract
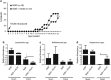
Accumulating evidence supports that the intestinal microbiome is involved in Type 1 diabetes (T1D) pathogenesis through the gut-pancreas nexus. Our aim was to determine whether the intestinal microbiota in the non-obese diabetic (NOD) mouse model played a role in T1D through the gut. To examine the effect of the intestinal microbiota on T1D onset, we manipulated gut microbes by: (1) the fecal transplantation between non-obese diabetic (NOD) and resistant (NOR) mice and (2) the oral antibiotic and probiotic treatment of NOD mice. We monitored diabetes onset, quantified CD4+T cells in the Peyer's patches, profiled the microbiome and measured fecal short-chain fatty acids (SCFA). The gut microbiota from NOD mice harbored more pathobionts and fewer beneficial microbes in comparison with NOR mice. Fecal transplantation of NOD microbes induced insulitis in NOR hosts suggesting that the NOD microbiome is diabetogenic. Moreover, antibiotic exposure accelerated diabetes onset in NOD mice accompanied by increased T-helper type 1 (Th1) and reduced Th17 cells in the intestinal lymphoid tissues. The diabetogenic microbiome was characterized by a metagenome altered in several metabolic gene clusters. Furthermore, diabetes susceptibility correlated with reduced fecal SCFAs. In an attempt to correct the diabetogenic microbiome, we administered VLS#3 probiotics to NOD mice but found that VSL#3 colonized the intestine poorly and did not delay diabetes. We conclude that NOD mice harbor gut microbes that induce diabetes and that their diabetogenic microbiome can be amplified early in life through antibiotic exposure. Protective microbes like VSL#3 are insufficient to overcome the effects of a diabetogenic microbiome.
Figures

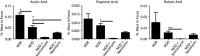
Similar articles
-
Antibiotic-associated Manipulation of the Gut Microbiota and Phenotypic Restoration in NOD Mice.Comp Med. 2017 Aug 1;67(4):335-343. Comp Med. 2017. PMID: 28830580 Free PMC article.
-
Oral Probiotic VSL#3 Prevents Autoimmune Diabetes by Modulating Microbiota and Promoting Indoleamine 2,3-Dioxygenase-Enriched Tolerogenic Intestinal Environment.J Diabetes Res. 2016;2016:7569431. doi: 10.1155/2016/7569431. Epub 2015 Dec 8. J Diabetes Res. 2016. PMID: 26779542 Free PMC article.
-
Modulation of Gut Microbiota by Low Methoxyl Pectin Attenuates Type 1 Diabetes in Non-obese Diabetic Mice.Front Immunol. 2019 Jul 30;10:1733. doi: 10.3389/fimmu.2019.01733. eCollection 2019. Front Immunol. 2019. PMID: 31417546 Free PMC article.
-
Th17 Cells in Type 1 Diabetes: Role in the Pathogenesis and Regulation by Gut Microbiome.Mediators Inflamm. 2015;2015:638470. doi: 10.1155/2015/638470. Epub 2015 Dec 30. Mediators Inflamm. 2015. PMID: 26843788 Free PMC article. Review.
-
The gut microbiota and Type 1 Diabetes.Clin Immunol. 2015 Aug;159(2):143-53. doi: 10.1016/j.clim.2015.05.013. Epub 2015 Jun 4. Clin Immunol. 2015. PMID: 26051037 Free PMC article. Review.
Cited by
-
The comprehensive evaluation of oral and fecal microbiota in patients with acromegaly.Pituitary. 2024 Oct;27(5):555-566. doi: 10.1007/s11102-024-01444-6. Epub 2024 Aug 19. Pituitary. 2024. PMID: 39158810
-
Microbiome bacterial influencers of host immunity and response to immunotherapy.Cell Rep Med. 2024 Apr 16;5(4):101487. doi: 10.1016/j.xcrm.2024.101487. Epub 2024 Mar 27. Cell Rep Med. 2024. PMID: 38547865 Free PMC article. Review.
-
Unlocking the secrets: exploring the influence of the aryl hydrocarbon receptor and microbiome on cancer development.Cell Mol Biol Lett. 2024 Mar 6;29(1):33. doi: 10.1186/s11658-024-00538-0. Cell Mol Biol Lett. 2024. PMID: 38448800 Free PMC article. Review.
-
Depletion of gut microbiota facilitates fibroblast growth factor 21-mediated protection against acute pancreatitis in diabetic mice.World J Diabetes. 2023 Dec 15;14(12):1824-1838. doi: 10.4239/wjd.v14.i12.1824. World J Diabetes. 2023. PMID: 38222783 Free PMC article.
-
The potential mechanism of gut microbiota-microbial metabolites-mitochondrial axis in progression of diabetic kidney disease.Mol Med. 2023 Oct 31;29(1):148. doi: 10.1186/s10020-023-00745-z. Mol Med. 2023. PMID: 37907885 Free PMC article. Review.
References
-
- Akerblom HK, Knip M. (1998). Putative environmental factors in Type 1 diabetes. Diabetes Metab Rev 14: 31–67. - PubMed
-
- Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. (2010). Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta 1801: 1175–1183. - PubMed
-
- Alam C, Bittoun E, Bhagwat D, Valkonen S, Saari A, Jaakkola U et al. (2011). Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia 54: 1398–1406. - PubMed
-
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
-
- Bray JR, Curtis JT. (1957). An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol Monogr 27: 326–349.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

