NEDD8 Ultimate Buster 1 Long (NUB1L) Protein Suppresses Atypical Neddylation and Promotes the Proteasomal Degradation of Misfolded Proteins
- PMID: 26260793
- PMCID: PMC4583023
- DOI: 10.1074/jbc.M115.664375
NEDD8 Ultimate Buster 1 Long (NUB1L) Protein Suppresses Atypical Neddylation and Promotes the Proteasomal Degradation of Misfolded Proteins
Abstract
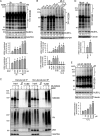
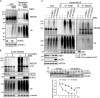
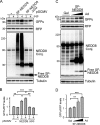
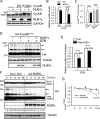
Neddylation is a posttranslational modification that controls diverse biological processes by covalently conjugating the ubiquitin-like protein NEDD8 to specific targets. Neddylation is commonly mediated by NEDD8-specific enzymes (typical neddylation) and, sometimes, by ubiquitin enzymes (atypical neddylation). Although typical neddylation is known to regulate protein function in many ways, the regulatory mechanisms and biological consequence of atypical neddylation remain largely unexplored. Here we report that NEDD8 conjugates were accumulated in the diseased hearts from mouse models and human patients. Proteotoxic stresses induced typical and atypical neddylation in cardiomyocytes. Loss of NUB1L exaggerated atypical neddylation, whereas NUB1L overexpression repressed atypical neddylation through promoting the degradation of NEDD8. Activation of atypical neddylation accumulated a surrogate misfolded protein, GFPu. In contrast, suppression of atypical neddylation by NUB1L overexpression enhanced GFPu degradation. Moreover, NUB1L depletion accumulated a cardiomyopathy-linked misfolded protein, CryAB(R120G), whereas NUB1L overexpression promoted its degradation through suppressing neddylation of ubiquitinated proteins in cardiomyocytes. Consequently, NUB1L protected cells from proteotoxic stress-induced cell injury. In summary, these data indicate that NUB1L suppresses atypical neddylation and promotes the degradation of misfolded proteins by the proteasome. Our findings also suggest that induction of NUB1L could potentially become a novel therapeutic strategy for diseases with increased proteotoxic stress.
Keywords: NEDD8; NUB1L; cardiomyopathy; posttranslational modification (PTM); proteasome; protein degradation; ubiquitin.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
NEDD8 ultimate buster-1 long (NUB1L) protein promotes transfer of NEDD8 to proteasome for degradation through the P97UFD1/NPL4 complex.J Biol Chem. 2013 Oct 25;288(43):31339-49. doi: 10.1074/jbc.M113.484816. Epub 2013 Sep 9. J Biol Chem. 2013. PMID: 24019527 Free PMC article.
-
NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation.J Biol Chem. 2004 Apr 16;279(16):16503-10. doi: 10.1074/jbc.M310114200. Epub 2004 Feb 2. J Biol Chem. 2004. PMID: 14757770
-
FAT10 and NUB1L bind to the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis.Nat Commun. 2012 Mar 20;3:749. doi: 10.1038/ncomms1752. Nat Commun. 2012. PMID: 22434192
-
Neddylation dysfunction in Alzheimer's disease.J Cell Mol Med. 2012 Nov;16(11):2583-91. doi: 10.1111/j.1582-4934.2012.01604.x. J Cell Mol Med. 2012. PMID: 22805479 Free PMC article. Review.
-
Protein neddylation: beyond cullin-RING ligases.Nat Rev Mol Cell Biol. 2015 Jan;16(1):30-44. doi: 10.1038/nrm3919. Nat Rev Mol Cell Biol. 2015. PMID: 25531226 Free PMC article. Review.
Cited by
-
Regulation of NUB1 Activity through Non-Proteolytic Mdm2-Mediated Ubiquitination.PLoS One. 2017 Jan 18;12(1):e0169988. doi: 10.1371/journal.pone.0169988. eCollection 2017. PLoS One. 2017. PMID: 28099510 Free PMC article.
-
Stress-induced NEDDylation promotes cytosolic protein aggregation through HDAC6 in a p62-dependent manner.iScience. 2021 Feb 6;24(3):102146. doi: 10.1016/j.isci.2021.102146. eCollection 2021 Mar 19. iScience. 2021. PMID: 33665565 Free PMC article.
-
Non-Proteasomal UbL-UbA Family of Proteins in Neurodegeneration.Int J Mol Sci. 2019 Apr 17;20(8):1893. doi: 10.3390/ijms20081893. Int J Mol Sci. 2019. PMID: 30999567 Free PMC article. Review.
-
HYPK coordinates degradation of polyneddylated proteins by autophagy.Autophagy. 2022 Aug;18(8):1763-1784. doi: 10.1080/15548627.2021.1997053. Epub 2021 Nov 26. Autophagy. 2022. PMID: 34836490 Free PMC article.
-
Dysfunctional Mitochondrial Dynamic and Oxidative Phosphorylation Precedes Cardiac Dysfunction in R120G-αB-Crystallin-Induced Desmin-Related Cardiomyopathy.J Am Heart Assoc. 2020 Dec;9(23):e017195. doi: 10.1161/JAHA.120.017195. Epub 2020 Nov 19. J Am Heart Assoc. 2020. PMID: 33208022 Free PMC article.
References
-
- Herrmann J., Lerman L. O., Lerman A. (2007) Ubiquitin and ubiquitin-like proteins in protein regulation. Circ. Res. 100, 1276–1291 - PubMed
-
- Kumar S., Yoshida Y., Noda M. (1993) Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem. Biophys. Res. Commun. 195, 393–399 - PubMed
-
- Watson I. R., Irwin M. S., Ohh M. (2011) NEDD8 pathways in cancer, sine quibus non. Cancer Cell 19, 168–176 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01HL124248/HL/NHLBI NIH HHS/United States
- R01 HL111480/HL/NHLBI NIH HHS/United States
- R01HL072166/HL/NHLBI NIH HHS/United States
- R01 HL124248/HL/NHLBI NIH HHS/United States
- R01 HL085629/HL/NHLBI NIH HHS/United States
- R01HL085629/HL/NHLBI NIH HHS/United States
- 14SDG18970040/AHA/American Heart Association-American Stroke Association/United States
- R01HL124251/HL/NHLBI NIH HHS/United States
- R01HL111480/HL/NHLBI NIH HHS/United States
- R01 HL072166/HL/NHLBI NIH HHS/United States
- R01 HL124251/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

