Next-generation sequencing to guide cancer therapy
- PMID: 26221189
- PMCID: PMC4517547
- DOI: 10.1186/s13073-015-0203-x
Next-generation sequencing to guide cancer therapy
Abstract
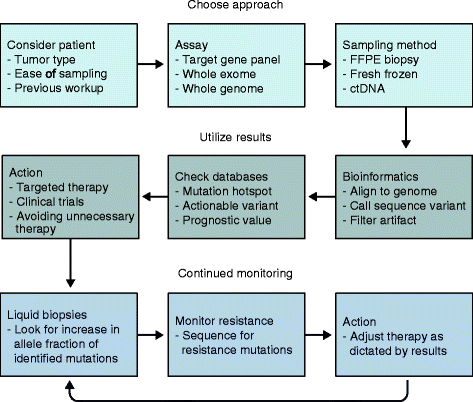
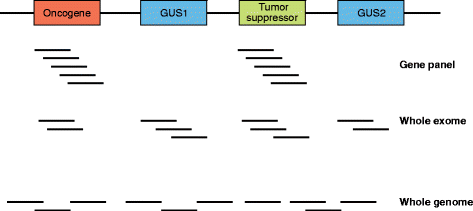
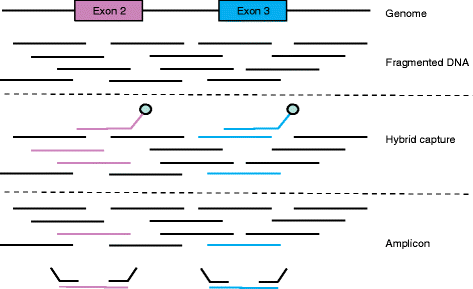
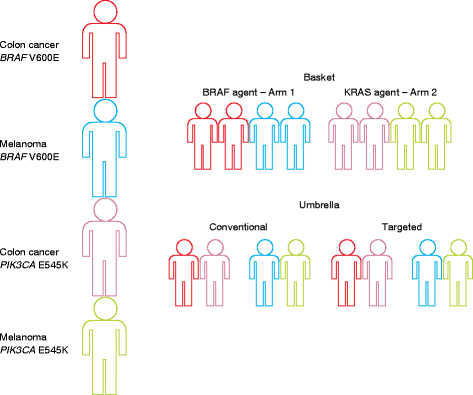
As a result of multiple technological and practical advances, high-throughput sequencing, known more commonly as "next-generation" sequencing (NGS), can now be incorporated into standard clinical practice. Whereas early protocols relied on samples that were harvested outside of typical clinical pathology workflows, standard formalin-fixed, paraffin-embedded specimens can more regularly be used as starting materials for NGS. Furthermore, protocols for the analysis and interpretation of NGS data, as well as knowledge bases, are being amassed, allowing clinicians to act more easily on genomic information at the point of care for patients. In parallel, new therapies that target somatically mutated genes identified through clinical NGS are gaining US Food and Drug Administration (FDA) approval, and novel clinical trial designs are emerging in which genetic identifiers are given equal weight to histology. For clinical oncology providers, understanding the potential and the limitations of DNA sequencing will be crucial for providing genomically driven care in this era of precision medicine.
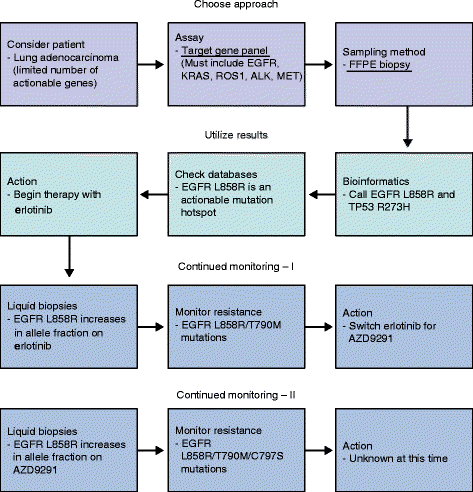
Figures
Similar articles
-
Comprehensive next-generation cancer genome sequencing in the era of targeted therapy and personalized oncology.Biomark Med. 2011 Jun;5(3):293-305. doi: 10.2217/bmm.11.37. Biomark Med. 2011. PMID: 21657839 Review.
-
A 2015 update on predictive molecular pathology and its role in targeted cancer therapy: a review focussing on clinical relevance.Cancer Gene Ther. 2015 Sep;22(9):417-30. doi: 10.1038/cgt.2015.39. Epub 2015 Sep 11. Cancer Gene Ther. 2015. PMID: 26358176 Review.
-
Targeted Next Generation Sequencing as a Reliable Diagnostic Assay for the Detection of Somatic Mutations in Tumours Using Minimal DNA Amounts from Formalin Fixed Paraffin Embedded Material.PLoS One. 2016 Feb 26;11(2):e0149405. doi: 10.1371/journal.pone.0149405. eCollection 2016. PLoS One. 2016. PMID: 26919633 Free PMC article.
-
Clinical next-generation sequencing in patients with non-small cell lung cancer.Cancer. 2015 Feb 15;121(4):631-9. doi: 10.1002/cncr.29089. Epub 2014 Oct 24. Cancer. 2015. PMID: 25345567
-
Personalized cancer therapy-leveraging a knowledge base for clinical decision-making.Cold Spring Harb Mol Case Stud. 2018 Apr 2;4(2):a001578. doi: 10.1101/mcs.a001578. Print 2018 Apr. Cold Spring Harb Mol Case Stud. 2018. PMID: 29212833 Free PMC article. Review.
Cited by
-
Genomic sequencing in clinical practice: applications, challenges, and opportunities.Dialogues Clin Neurosci. 2016 Sep;18(3):299-312. doi: 10.31887/DCNS.2016.18.3/jkrier. Dialogues Clin Neurosci. 2016. PMID: 27757064 Free PMC article. Review.
-
An analysis of the relationship between metastases and cachexia in lung cancer patients.Cancer Med. 2016 Sep;5(9):2641-8. doi: 10.1002/cam4.841. Epub 2016 Aug 3. Cancer Med. 2016. PMID: 27485414 Free PMC article.
-
Antibody-Drug Conjugates and Their Potential in the Treatment of Patients with Biliary Tract Cancer.Cancers (Basel). 2024 Sep 30;16(19):3345. doi: 10.3390/cancers16193345. Cancers (Basel). 2024. PMID: 39409965 Free PMC article. Review.
-
Clinical target sequencing for precision medicine of breast cancer.Int J Clin Oncol. 2019 Feb;24(2):131-140. doi: 10.1007/s10147-018-1373-5. Epub 2019 Jan 2. Int J Clin Oncol. 2019. PMID: 30604156 Review.
-
Comprehensive analysis of genomic alterations detected by next-generation sequencing-based tissue and circulating tumor DNA assays in Chinese patients with non-small cell lung cancer.Oncol Lett. 2019 Nov;18(5):4762-4770. doi: 10.3892/ol.2019.10791. Epub 2019 Sep 3. Oncol Lett. 2019. PMID: 31611986 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

