Curcumin-Mediated HDAC Inhibition Suppresses the DNA Damage Response and Contributes to Increased DNA Damage Sensitivity
- PMID: 26218133
- PMCID: PMC4517890
- DOI: 10.1371/journal.pone.0134110
Curcumin-Mediated HDAC Inhibition Suppresses the DNA Damage Response and Contributes to Increased DNA Damage Sensitivity
Abstract
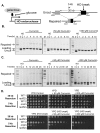
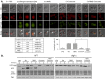
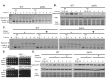
Chemo- and radiotherapy cause multiple forms of DNA damage and lead to the death of cancer cells. Inhibitors of the DNA damage response are candidate drugs for use in combination therapies to increase the efficacy of such treatments. In this study, we show that curcumin, a plant polyphenol, sensitizes budding yeast to DNA damage by counteracting the DNA damage response. Following DNA damage, the Mec1-dependent DNA damage checkpoint is inactivated and Rad52 recombinase is degraded by curcumin, which results in deficiencies in double-stand break repair. Additive effects on damage-induced apoptosis and the inhibition of damage-induced autophagy by curcumin were observed. Moreover, rpd3 mutants were found to mimic the curcumin-induced suppression of the DNA damage response. In contrast, hat1 mutants were resistant to DNA damage, and Rad52 degradation was impaired following curcumin treatment. These results indicate that the histone deacetylase inhibitor activity of curcumin is critical to DSB repair and DNA damage sensitivity.
Conflict of interest statement
Figures




Similar articles
-
Curcumin suppresses multiple DNA damage response pathways and has potency as a sensitizer to PARP inhibitor.Carcinogenesis. 2013 Nov;34(11):2486-97. doi: 10.1093/carcin/bgt240. Epub 2013 Jul 3. Carcinogenesis. 2013. PMID: 23825154
-
Dihydrocoumarin, an HDAC Inhibitor, Increases DNA Damage Sensitivity by Inhibiting Rad52.Int J Mol Sci. 2017 Dec 7;18(12):2655. doi: 10.3390/ijms18122655. Int J Mol Sci. 2017. PMID: 29215575 Free PMC article.
-
HDACs link the DNA damage response, processing of double-strand breaks and autophagy.Nature. 2011 Mar 3;471(7336):74-79. doi: 10.1038/nature09803. Nature. 2011. PMID: 21368826 Free PMC article.
-
Histone Deacetylase Inhibitors as Anticancer Drugs.Int J Mol Sci. 2017 Jul 1;18(7):1414. doi: 10.3390/ijms18071414. Int J Mol Sci. 2017. PMID: 28671573 Free PMC article. Review.
-
HDAC inhibitors: roles of DNA damage and repair.Adv Cancer Res. 2012;116:87-129. doi: 10.1016/B978-0-12-394387-3.00003-3. Adv Cancer Res. 2012. PMID: 23088869 Review.
Cited by
-
Genome-Protecting Compounds as Potential Geroprotectors.Int J Mol Sci. 2020 Jun 24;21(12):4484. doi: 10.3390/ijms21124484. Int J Mol Sci. 2020. PMID: 32599754 Free PMC article. Review.
-
The Essential Medicinal Chemistry of Curcumin.J Med Chem. 2017 Mar 9;60(5):1620-1637. doi: 10.1021/acs.jmedchem.6b00975. Epub 2017 Jan 11. J Med Chem. 2017. PMID: 28074653 Free PMC article. Review.
-
Emerging Therapeutic Targets Against Toxoplasma gondii: Update on DNA Repair Response Inhibitors and Genotoxic Drugs.Front Cell Infect Microbiol. 2020 Jun 12;10:289. doi: 10.3389/fcimb.2020.00289. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32656097 Free PMC article. Review.
-
Genetic interactions derived from high-throughput phenotyping of 6589 yeast cell cycle mutants.NPJ Syst Biol Appl. 2020 May 6;6(1):11. doi: 10.1038/s41540-020-0134-z. NPJ Syst Biol Appl. 2020. PMID: 32376972 Free PMC article.
-
Novel Insights into RAD52's Structure, Function, and Druggability for Synthetic Lethality and Innovative Anticancer Therapies.Cancers (Basel). 2023 Mar 17;15(6):1817. doi: 10.3390/cancers15061817. Cancers (Basel). 2023. PMID: 36980703 Free PMC article. Review.
References
-
- Qin J, Li L. Molecular anatomy of the DNA damage and replication checkpoints. Radiation research. 2003;159(2):139–48. Epub 2003/01/23. . - PubMed
-
- Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Current opinion in cell biology. 2002;14(2):237–45. Epub 2002/03/14. . - PubMed
-
- Gilbert CS, Green CM, Lowndes NF. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell. 2001;8(1):129–36. Epub 2001/08/21. . - PubMed
-
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271(5247):357–60. Epub 1996/01/19. . - PubMed
-
- Dietlein F, Reinhardt HC. Molecular pathways: exploiting tumor-specific molecular defects in DNA repair pathways for precision cancer therapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(23):5882–7. Epub 2014/12/03. 10.1158/1078-0432.CCR-14-1165 . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

