Chlamydia trachomatis Type III Secretion Proteins Regulate Transcription
- PMID: 26216849
- PMCID: PMC4573728
- DOI: 10.1128/JB.00379-15
Chlamydia trachomatis Type III Secretion Proteins Regulate Transcription
Abstract
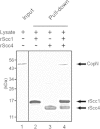
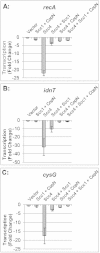
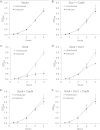
The Scc4 protein (CT663) of the pathogenic bacterium Chlamydia has been described as a type III secretion (T3S) chaperone as well as an inhibitor of RNA polymerase. To examine if these roles are connected, we first investigated physical interactions between Chlamydia trachomatis Scc4 and the T3S chaperone Scc1 and a T3S substrate, CopN. In a yeast 3-hybrid assay, Scc4, Scc1, and CopN were all required to detect an interaction, which suggests that these proteins form a trimolecular complex. We also detected interactions between any two of these three T3S proteins in a pulldown assay using only recombinant proteins. We next determined whether these interactions affected the function of Scc4 as an inhibitor of RNA transcription. Using Escherichia coli as a heterologous in vivo system, we demonstrated that expression of C. trachomatis Scc4 led to a drastic decrease in transcript levels for multiple genes. However, coexpression of Scc4 with Scc1, CopN, or both alleviated Scc4-mediated inhibition of transcription. Scc4 expression also severely impaired E. coli growth, but this growth defect was reversed by coexpression of Scc4 with Scc1, CopN, or both, suggesting that the inhibitory effect of Scc4 on transcription and growth can be antagonized by interactions between Scc4, Scc1, and CopN. These findings suggest that the dual functions of Scc4 may serve as a bridge to link T3S and the regulation of gene expression in Chlamydia.
Importance: This study investigates a novel mechanism for regulating gene expression in the pathogenic bacterium Chlamydia. The Chlamydia type III secretion (T3S) chaperone Scc4 has been shown to inhibit transcription by RNA polymerase. This study describes physical interactions between Scc4 and the T3S proteins Scc1 and CopN. Furthermore, Chlamydia Scc1 and CopN antagonized the inhibitory effects of Scc4 on transcription and growth in a heterologous Escherichia coli system. These results provide evidence that transcription in Chlamydia can be regulated by the T3S system through interactions between T3S proteins.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Context-Dependent Action of Scc4 Reinforces Control of the Type III Secretion System.J Bacteriol. 2020 Jul 9;202(15):e00132-20. doi: 10.1128/JB.00132-20. Print 2020 Jul 9. J Bacteriol. 2020. PMID: 32424009 Free PMC article.
-
Multipart Chaperone-Effector Recognition in the Type III Secretion System of Chlamydia trachomatis.J Biol Chem. 2015 Nov 20;290(47):28141-28155. doi: 10.1074/jbc.M115.670232. Epub 2015 Oct 5. J Biol Chem. 2015. PMID: 26438824 Free PMC article.
-
Insights into the association of the Chlamydia trachomatis type III secretion chaperone complex, Scc4:Scc1, from sequential expression in Escherichia coli.Protein Expr Purif. 2024 Oct;222:106532. doi: 10.1016/j.pep.2024.106532. Epub 2024 Jun 8. Protein Expr Purif. 2024. PMID: 38857716
-
Sigma and RNA polymerase: an on-again, off-again relationship?Mol Cell. 2005 Nov 11;20(3):335-45. doi: 10.1016/j.molcel.2005.10.015. Mol Cell. 2005. PMID: 16285916 Review.
-
Evolutionary Conservation, Variability, and Adaptation of Type III Secretion Systems.J Membr Biol. 2022 Oct;255(4-5):599-612. doi: 10.1007/s00232-022-00247-9. Epub 2022 Jun 13. J Membr Biol. 2022. PMID: 35695900 Review.
Cited by
-
A working model for the type III secretion mechanism in Chlamydia.Microbes Infect. 2016 Feb;18(2):84-92. doi: 10.1016/j.micinf.2015.10.006. Epub 2015 Oct 26. Microbes Infect. 2016. PMID: 26515030 Free PMC article. Review.
-
Type III Secretion in Chlamydia.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0003423. doi: 10.1128/mmbr.00034-23. Epub 2023 Jun 26. Microbiol Mol Biol Rev. 2023. PMID: 37358451 Free PMC article. Review.
-
Chlamydia cell biology and pathogenesis.Nat Rev Microbiol. 2016 Jun;14(6):385-400. doi: 10.1038/nrmicro.2016.30. Epub 2016 Apr 25. Nat Rev Microbiol. 2016. PMID: 27108705 Free PMC article. Review.
-
EspM Is a Conserved Transcription Factor That Regulates Gene Expression in Response to the ESX-1 System.mBio. 2020 Feb 4;11(1):e02807-19. doi: 10.1128/mBio.02807-19. mBio. 2020. PMID: 32019792 Free PMC article.
-
MxiA, MxiC and IpaD Regulate Substrate Selection and Secretion Mode in the T3SS of Shigella flexneri.PLoS One. 2016 May 12;11(5):e0155141. doi: 10.1371/journal.pone.0155141. eCollection 2016. PLoS One. 2016. PMID: 27171191 Free PMC article.
References
-
- Batteiger BE, Tan M. 2014. Chlamydia trachomatis (trachoma, genital infections, perinatal infections, and lymphogranuloma venereum), p 2154–2170. In Bennett JE, Dolin R, Mandell GL (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 8th ed Elsevier Inc, Philadelphia, PA.
-
- Adams DA, Jajosky RA, Ajani U, Kriseman J, Sharp P, Onwen DH, Schley AW, Anderson WJ, Grigoryan A, Aranas AE, Wodajo MS, Abellera JP, Centers for Disease Control and Prevention (CDC). 2014. Summary of notifiable diseases—United States, 2012. MMWR Morb Mortal Wkly Rep 61:1–121. - PubMed

