Genomics and drug profiling of fatal TCF3-HLF-positive acute lymphoblastic leukemia identifies recurrent mutation patterns and therapeutic options
- PMID: 26214592
- PMCID: PMC4603357
- DOI: 10.1038/ng.3362
Genomics and drug profiling of fatal TCF3-HLF-positive acute lymphoblastic leukemia identifies recurrent mutation patterns and therapeutic options
Abstract
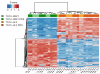
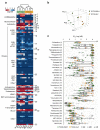
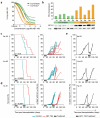
TCF3-HLF-positive acute lymphoblastic leukemia (ALL) is currently incurable. Using an integrated approach, we uncovered distinct mutation, gene expression and drug response profiles in TCF3-HLF-positive and treatment-responsive TCF3-PBX1-positive ALL. We identified recurrent intragenic deletions of PAX5 or VPREB1 in constellation with the fusion of TCF3 and HLF. Moreover somatic mutations in the non-translocated allele of TCF3 and a reduction of PAX5 gene dosage in TCF3-HLF ALL suggest cooperation within a restricted genetic context. The enrichment for stem cell and myeloid features in the TCF3-HLF signature may reflect reprogramming by TCF3-HLF of a lymphoid-committed cell of origin toward a hybrid, drug-resistant hematopoietic state. Drug response profiling of matched patient-derived xenografts revealed a distinct profile for TCF3-HLF ALL with resistance to conventional chemotherapeutics but sensitivity to glucocorticoids, anthracyclines and agents in clinical development. Striking on-target sensitivity was achieved with the BCL2-specific inhibitor venetoclax (ABT-199). This integrated approach thus provides alternative treatment options for this deadly disease.
Figures



Comment in
-
Haematological cancer: promising results of BCL2 inhibition.Nat Rev Clin Oncol. 2015 Sep;12(9):504. doi: 10.1038/nrclinonc.2015.139. Epub 2015 Aug 11. Nat Rev Clin Oncol. 2015. PMID: 26260042 No abstract available.
Similar articles
-
The Leukemogenic TCF3-HLF Complex Rewires Enhancers Driving Cellular Identity and Self-Renewal Conferring EP300 Vulnerability.Cancer Cell. 2019 Dec 9;36(6):630-644.e9. doi: 10.1016/j.ccell.2019.10.004. Epub 2019 Nov 14. Cancer Cell. 2019. PMID: 31735627
-
Comprehensive chromosomal aberrations in a case of a patient with TCF3-HLF-positive BCP-ALL.BMC Med Genomics. 2020 Apr 3;13(1):58. doi: 10.1186/s12920-020-0709-y. BMC Med Genomics. 2020. PMID: 32245383 Free PMC article.
-
NGS-based methylation profiling differentiates TCF3-HLF and TCF3-PBX1 positive B-cell acute lymphoblastic leukemia.Epigenomics. 2018 Feb;10(2):133-147. doi: 10.2217/epi-2017-0080. Epub 2018 Jan 15. Epigenomics. 2018. PMID: 29334255
-
PAX5 fusion genes in acute lymphoblastic leukemia: A literature review.Medicine (Baltimore). 2023 May 19;102(20):e33836. doi: 10.1097/MD.0000000000033836. Medicine (Baltimore). 2023. PMID: 37335685 Free PMC article. Review.
-
Regulation of the miRNA expression by TEL/AML1, BCR/ABL, MLL/AF4 and TCF3/PBX1 oncoproteins in acute lymphoblastic leukemia (Review).Oncol Rep. 2016 Sep;36(3):1226-32. doi: 10.3892/or.2016.4948. Epub 2016 Jul 19. Oncol Rep. 2016. PMID: 27431573 Review.
Cited by
-
CBP Modulates Sensitivity to Dasatinib in Pre-BCR+ Acute Lymphoblastic Leukemia.Cancer Res. 2018 Nov 15;78(22):6497-6508. doi: 10.1158/0008-5472.CAN-18-1703. Epub 2018 Sep 27. Cancer Res. 2018. PMID: 30262461 Free PMC article.
-
International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data.Blood. 2022 Sep 15;140(11):1200-1228. doi: 10.1182/blood.2022015850. Blood. 2022. PMID: 35767897 Free PMC article.
-
Engineering of CD19 Antibodies: A CD19-TRAIL Fusion Construct Specifically Induces Apoptosis in B-Cell Precursor Acute Lymphoblastic Leukemia (BCP-ALL) Cells In Vivo.J Clin Med. 2021 Jun 15;10(12):2634. doi: 10.3390/jcm10122634. J Clin Med. 2021. PMID: 34203833 Free PMC article.
-
The Clinical Utility of Optical Genome Mapping for the Assessment of Genomic Aberrations in Acute Lymphoblastic Leukemia.Cancers (Basel). 2021 Aug 30;13(17):4388. doi: 10.3390/cancers13174388. Cancers (Basel). 2021. PMID: 34503197 Free PMC article.
-
An in-depth analysis reveals two new genetic variants on 22q11.2 associated with vitiligo in the Chinese Han population.Mol Biol Rep. 2021 Aug;48(8):5955-5964. doi: 10.1007/s11033-021-06597-2. Epub 2021 Aug 4. Mol Biol Rep. 2021. PMID: 34350550
References
-
- Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. - PubMed
-
- Russell LJ, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009;114:2688–2698. - PubMed
-
- Hertzberg L, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115:1006–1017. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

