miR-302 Is Required for Timing of Neural Differentiation, Neural Tube Closure, and Embryonic Viability
- PMID: 26212322
- PMCID: PMC4741278
- DOI: 10.1016/j.celrep.2015.06.074
miR-302 Is Required for Timing of Neural Differentiation, Neural Tube Closure, and Embryonic Viability
Abstract
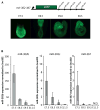
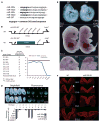
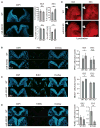
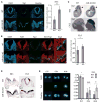
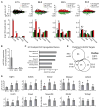
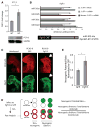
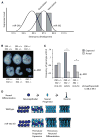
The evolutionarily conserved miR-302 family of microRNAs is expressed during early mammalian embryonic development. Here, we report that deletion of miR-302a-d in mice results in a fully penetrant late embryonic lethal phenotype. Knockout embryos have an anterior neural tube closure defect associated with a thickened neuroepithelium. The neuroepithelium shows increased progenitor proliferation, decreased cell death, and precocious neuronal differentiation. mRNA profiling at multiple time points during neurulation uncovers a complex pattern of changing targets over time. Overexpression of one of these targets, Fgf15, in the neuroepithelium of the chick embryo induces precocious neuronal differentiation. Compound mutants between mir-302 and the related mir-290 locus have a synthetic lethal phenotype prior to neurulation. Our results show that mir-302 helps regulate neurulation by suppressing neural progenitor expansion and precocious differentiation. Furthermore, these results uncover redundant roles for mir-290 and mir-302 early in development.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
MiR-302/367 regulate neural progenitor proliferation, differentiation timing, and survival in neurulation.Dev Biol. 2015 Dec 1;408(1):140-50. doi: 10.1016/j.ydbio.2015.09.020. Epub 2015 Oct 9. Dev Biol. 2015. PMID: 26441343 Free PMC article.
-
Bioinformatics and microarray analysis of microRNA expression profiles of murine embryonic stem cells, neural stem cells induced from ESCs and isolated from E8.5 mouse neural tube.Neurol Res. 2010 Jul;32(6):603-13. doi: 10.1179/174313209X455691. Epub 2009 Aug 5. Neurol Res. 2010. PMID: 19660235
-
MiR-302 Regulates Glycolysis to Control Cell-Cycle during Neural Tube Closure.Int J Mol Sci. 2020 Oct 13;21(20):7534. doi: 10.3390/ijms21207534. Int J Mol Sci. 2020. PMID: 33066028 Free PMC article.
-
In vitro recapitulation of neural development using embryonic stem cells: from neurogenesis to histogenesis.Dev Growth Differ. 2012 Apr;54(3):349-57. doi: 10.1111/j.1440-169X.2012.01329.x. Epub 2012 Mar 8. Dev Growth Differ. 2012. PMID: 22404483 Review.
-
[Neurulation continues: the parade commander is...apical constriction].Ontogenez. 2014 Jul-Aug;45(4):240-9. Ontogenez. 2014. PMID: 25735147 Review. Russian.
Cited by
-
Roles of MicroRNAs in Establishing and Modulating Stem Cell Potential.Int J Mol Sci. 2019 Jul 25;20(15):3643. doi: 10.3390/ijms20153643. Int J Mol Sci. 2019. PMID: 31349654 Free PMC article. Review.
-
p53 Mutant p53N236S Induces Neural Tube Defects in Female Embryos.Int J Biol Sci. 2019 Jul 25;15(9):2006-2015. doi: 10.7150/ijbs.31451. eCollection 2019. Int J Biol Sci. 2019. PMID: 31523200 Free PMC article.
-
Regulation of Oct4 in stem cells and neural crest cells.Birth Defects Res. 2022 Oct 1;114(16):983-1002. doi: 10.1002/bdr2.2007. Epub 2022 Apr 1. Birth Defects Res. 2022. PMID: 35365980 Free PMC article. Review.
-
LIS1 RNA-binding orchestrates the mechanosensitive properties of embryonic stem cells in AGO2-dependent and independent ways.Nat Commun. 2023 Jun 6;14(1):3293. doi: 10.1038/s41467-023-38797-8. Nat Commun. 2023. PMID: 37280197 Free PMC article.
-
MicroRNAs Instruct and Maintain Cell Type Diversity in the Nervous System.Front Mol Neurosci. 2021 Apr 29;14:646072. doi: 10.3389/fnmol.2021.646072. eCollection 2021. Front Mol Neurosci. 2021. PMID: 33994943 Free PMC article. Review.
References
-
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

