Evolution of selenophosphate synthetases: emergence and relocation of function through independent duplications and recurrent subfunctionalization
- PMID: 26194102
- PMCID: PMC4561486
- DOI: 10.1101/gr.190538.115
Evolution of selenophosphate synthetases: emergence and relocation of function through independent duplications and recurrent subfunctionalization
Abstract
Selenoproteins are proteins that incorporate selenocysteine (Sec), a nonstandard amino acid encoded by UGA, normally a stop codon. Sec synthesis requires the enzyme Selenophosphate synthetase (SPS or SelD), conserved in all prokaryotic and eukaryotic genomes encoding selenoproteins. Here, we study the evolutionary history of SPS genes, providing a map of selenoprotein function spanning the whole tree of life. SPS is itself a selenoprotein in many species, although functionally equivalent homologs that replace the Sec site with cysteine (Cys) are common. Many metazoans, however, possess SPS genes with substitutions other than Sec or Cys (collectively referred to as SPS1). Using complementation assays in fly mutants, we show that these genes share a common function, which appears to be distinct from the synthesis of selenophosphate carried out by the Sec- and Cys- SPS genes (termed SPS2), and unrelated to Sec synthesis. We show here that SPS1 genes originated through a number of independent gene duplications from an ancestral metazoan selenoprotein SPS2 gene that most likely already carried the SPS1 function. Thus, in SPS genes, parallel duplications and subsequent convergent subfunctionalization have resulted in the segregation to different loci of functions initially carried by a single gene. This evolutionary history constitutes a remarkable example of emergence and evolution of gene function, which we have been able to trace thanks to the singular features of SPS genes, wherein the amino acid at a single site determines unequivocally protein function and is intertwined to the evolutionary fate of the entire selenoproteome.
© 2015 Mariotti et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
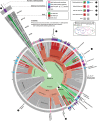
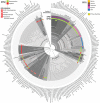


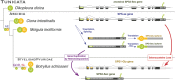

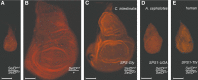
Similar articles
-
Asgard archaeal selenoproteome reveals a roadmap for the archaea-to-eukaryote transition of selenocysteine incorporation machinery.ISME J. 2024 Jan 8;18(1):wrae111. doi: 10.1093/ismejo/wrae111. ISME J. 2024. PMID: 38896033 Free PMC article.
-
The selenophosphate synthetase family: A review.Free Radic Biol Med. 2022 Nov 1;192:63-76. doi: 10.1016/j.freeradbiomed.2022.09.007. Epub 2022 Sep 16. Free Radic Biol Med. 2022. PMID: 36122644 Review.
-
Selenophosphate synthetase genes from lung adenocarcinoma cells: Sps1 for recycling L-selenocysteine and Sps2 for selenite assimilation.Proc Natl Acad Sci U S A. 2004 Nov 16;101(46):16162-7. doi: 10.1073/pnas.0406313101. Epub 2004 Nov 8. Proc Natl Acad Sci U S A. 2004. PMID: 15534230 Free PMC article.
-
Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis.Biochem J. 2007 May 15;404(1):115-20. doi: 10.1042/BJ20070165. Biochem J. 2007. PMID: 17346238 Free PMC article.
-
The selenium to selenoprotein pathway in eukaryotes: more molecular partners than anticipated.Biochim Biophys Acta. 2009 Nov;1790(11):1415-23. doi: 10.1016/j.bbagen.2009.03.003. Epub 2009 Mar 11. Biochim Biophys Acta. 2009. PMID: 19285539 Review.
Cited by
-
Time-Resolved Examination of Fungal Selenium Redox Transformations.ACS Earth Space Chem. 2023 May 5;7(5):960-971. doi: 10.1021/acsearthspacechem.2c00288. eCollection 2023 May 18. ACS Earth Space Chem. 2023. PMID: 37228623 Free PMC article.
-
Selenium Metabolism and Selenoproteins in Prokaryotes: A Bioinformatics Perspective.Biomolecules. 2022 Jun 29;12(7):917. doi: 10.3390/biom12070917. Biomolecules. 2022. PMID: 35883471 Free PMC article. Review.
-
Selenocysteine Machinery Primarily Supports TXNRD1 and GPX4 Functions and Together They Are Functionally Linked with SCD and PRDX6.Biomolecules. 2022 Jul 28;12(8):1049. doi: 10.3390/biom12081049. Biomolecules. 2022. PMID: 36008942 Free PMC article.
-
Asgard archaeal selenoproteome reveals a roadmap for the archaea-to-eukaryote transition of selenocysteine incorporation machinery.ISME J. 2024 Jan 8;18(1):wrae111. doi: 10.1093/ismejo/wrae111. ISME J. 2024. PMID: 38896033 Free PMC article.
-
Construction of anti-codon table of the plant kingdom and evolution of tRNA selenocysteine (tRNASec).BMC Genomics. 2020 Nov 19;21(1):804. doi: 10.1186/s12864-020-07216-3. BMC Genomics. 2020. PMID: 33213362 Free PMC article.
References
-
- Allmang C, Wurth L, Krol A. 2009. The selenium to selenoprotein pathway in eukaryotes: more molecular partners than anticipated. Biochim Biophys Acta 1790: 1415–1423. - PubMed
-
- Alsina B, Serras F, Baguñá J, Corominas M. 1998. patufet, the gene encoding the Drosophila melanogaster homologue of selenophosphate synthetase, is involved in imaginal disc morphogenesis. Mol Gen Genet 257: 113–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
