Identification of Spen as a Crucial Factor for Xist Function through Forward Genetic Screening in Haploid Embryonic Stem Cells
- PMID: 26190100
- PMCID: PMC4530576
- DOI: 10.1016/j.celrep.2015.06.067
Identification of Spen as a Crucial Factor for Xist Function through Forward Genetic Screening in Haploid Embryonic Stem Cells
Abstract
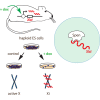
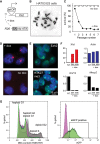
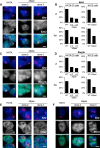
In mammals, the noncoding Xist RNA triggers transcriptional silencing of one of the two X chromosomes in female cells. Here, we report a genetic screen for silencing factors in X chromosome inactivation using haploid mouse embryonic stem cells (ESCs) that carry an engineered selectable reporter system. This system was able to identify several candidate factors that are genetically required for chromosomal repression by Xist. Among the list of candidates, we identify the RNA-binding protein Spen, the homolog of split ends. Independent validation through gene deletion in ESCs confirms that Spen is required for gene repression by Xist. However, Spen is not required for Xist RNA localization and the recruitment of chromatin modifications, including Polycomb protein Ezh2. The identification of Spen opens avenues for further investigation into the gene-silencing pathway of Xist and shows the usefulness of haploid ESCs for genetic screening of epigenetic pathways.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Spen links RNA-mediated endogenous retrovirus silencing and X chromosome inactivation.Elife. 2020 May 7;9:e54508. doi: 10.7554/eLife.54508. Elife. 2020. PMID: 32379046 Free PMC article.
-
SPEN integrates transcriptional and epigenetic control of X-inactivation.Nature. 2020 Feb;578(7795):455-460. doi: 10.1038/s41586-020-1974-9. Epub 2020 Feb 5. Nature. 2020. PMID: 32025035 Free PMC article.
-
A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing.Cell Rep. 2015 Jul 28;12(4):562-72. doi: 10.1016/j.celrep.2015.06.053. Epub 2015 Jul 16. Cell Rep. 2015. PMID: 26190105 Free PMC article.
-
Progress toward understanding chromosome silencing by Xist RNA.Genes Dev. 2020 Jun 1;34(11-12):733-744. doi: 10.1101/gad.337196.120. Genes Dev. 2020. PMID: 32482714 Free PMC article. Review.
-
The B-side of Xist.F1000Res. 2020 Jan 28;9:F1000 Faculty Rev-55. doi: 10.12688/f1000research.21362.1. eCollection 2020. F1000Res. 2020. PMID: 32047616 Free PMC article. Review.
Cited by
-
The interaction of PRC2 with RNA or chromatin is mutually antagonistic.Genome Res. 2016 Jul;26(7):896-907. doi: 10.1101/gr.197632.115. Epub 2016 May 9. Genome Res. 2016. PMID: 27197219 Free PMC article.
-
Gene regulation in time and space during X-chromosome inactivation.Nat Rev Mol Cell Biol. 2022 Apr;23(4):231-249. doi: 10.1038/s41580-021-00438-7. Epub 2022 Jan 10. Nat Rev Mol Cell Biol. 2022. PMID: 35013589 Review.
-
An Overview of Long Noncoding RNAs Involved in Bone Regeneration from Mesenchymal Stem Cells.Stem Cells Int. 2018 Jan 28;2018:8273648. doi: 10.1155/2018/8273648. eCollection 2018. Stem Cells Int. 2018. PMID: 29535782 Free PMC article. Review.
-
Ten principles of heterochromatin formation and function.Nat Rev Mol Cell Biol. 2018 Apr;19(4):229-244. doi: 10.1038/nrm.2017.119. Epub 2017 Dec 13. Nat Rev Mol Cell Biol. 2018. PMID: 29235574 Free PMC article. Review.
-
Polyploidy of semi-cloned embryos generated from parthenogenetic haploid embryonic stem cells.PLoS One. 2020 Sep 10;15(9):e0233072. doi: 10.1371/journal.pone.0233072. eCollection 2020. PLoS One. 2020. PMID: 32911495 Free PMC article.
References
-
- Blewitt M.E., Gendrel A.V., Pang Z., Sparrow D.B., Whitelaw N., Craig J.M., Apedaile A., Hilton D.J., Dunwoodie S.L., Brockdorff N. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 2008;40:663–669. - PubMed
-
- Borsani G., Tonlorenzi R., Simmler M.C., Dandolo L., Arnaud D., Capra V., Grompe M., Pizzuti A., Muzny D., Lawrence C. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. - PubMed
-
- Brockdorff N., Ashworth A., Kay G.F., Cooper P., Smith S., McCabe V.M., Norris D.P., Penny G.D., Patel D., Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

