The BioPlex Network: A Systematic Exploration of the Human Interactome
- PMID: 26186194
- PMCID: PMC4617211
- DOI: 10.1016/j.cell.2015.06.043
The BioPlex Network: A Systematic Exploration of the Human Interactome
Abstract
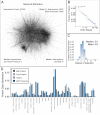
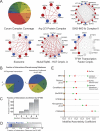
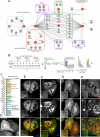
Protein interactions form a network whose structure drives cellular function and whose organization informs biological inquiry. Using high-throughput affinity-purification mass spectrometry, we identify interacting partners for 2,594 human proteins in HEK293T cells. The resulting network (BioPlex) contains 23,744 interactions among 7,668 proteins with 86% previously undocumented. BioPlex accurately depicts known complexes, attaining 80%-100% coverage for most CORUM complexes. The network readily subdivides into communities that correspond to complexes or clusters of functionally related proteins. More generally, network architecture reflects cellular localization, biological process, and molecular function, enabling functional characterization of thousands of proteins. Network structure also reveals associations among thousands of protein domains, suggesting a basis for examining structurally related proteins. Finally, BioPlex, in combination with other approaches, can be used to reveal interactions of biological or clinical significance. For example, mutations in the membrane protein VAPB implicated in familial amyotrophic lateral sclerosis perturb a defined community of interactors.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Architecture of the human interactome defines protein communities and disease networks.Nature. 2017 May 25;545(7655):505-509. doi: 10.1038/nature22366. Epub 2017 May 17. Nature. 2017. PMID: 28514442 Free PMC article.
-
Dual proteome-scale networks reveal cell-specific remodeling of the human interactome.Cell. 2021 May 27;184(11):3022-3040.e28. doi: 10.1016/j.cell.2021.04.011. Epub 2021 May 6. Cell. 2021. PMID: 33961781 Free PMC article.
-
BioPlex Display: An Interactive Suite for Large-Scale AP-MS Protein-Protein Interaction Data.J Proteome Res. 2018 Jan 5;17(1):722-726. doi: 10.1021/acs.jproteome.7b00572. Epub 2017 Oct 31. J Proteome Res. 2018. PMID: 29054129 Free PMC article.
-
Unraveling the dynamics of protein interactions with quantitative mass spectrometry.Crit Rev Biochem Mol Biol. 2011 Jun;46(3):216-28. doi: 10.3109/10409238.2011.567244. Epub 2011 Mar 26. Crit Rev Biochem Mol Biol. 2011. PMID: 21438726 Review.
-
Characterizing Protein-Protein Interactions Using Mass Spectrometry: Challenges and Opportunities.Trends Biotechnol. 2016 Oct;34(10):825-834. doi: 10.1016/j.tibtech.2016.02.014. Epub 2016 Mar 17. Trends Biotechnol. 2016. PMID: 26996615 Review.
Cited by
-
RTN4B-mediated suppression of Sirtuin 2 activity ameliorates β-amyloid pathology and cognitive impairment in Alzheimer's disease mouse model.Aging Cell. 2020 Aug;19(8):e13194. doi: 10.1111/acel.13194. Epub 2020 Jul 23. Aging Cell. 2020. PMID: 32700357 Free PMC article.
-
The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis.Elife. 2016 Aug 19;5:e17828. doi: 10.7554/eLife.17828. Elife. 2016. PMID: 27540631 Free PMC article.
-
Recent progress in mass spectrometry-based strategies for elucidating protein-protein interactions.Cell Mol Life Sci. 2021 Jul;78(13):5325-5339. doi: 10.1007/s00018-021-03856-0. Epub 2021 May 27. Cell Mol Life Sci. 2021. PMID: 34046695 Free PMC article. Review.
-
The SMN-ribosome interplay: a new opportunity for Spinal Muscular Atrophy therapies.Biochem Soc Trans. 2024 Feb 28;52(1):465-479. doi: 10.1042/BST20231116. Biochem Soc Trans. 2024. PMID: 38391004 Free PMC article. Review.
-
Target identification among known drugs by deep learning from heterogeneous networks.Chem Sci. 2020 Jan 13;11(7):1775-1797. doi: 10.1039/c9sc04336e. Chem Sci. 2020. PMID: 34123272 Free PMC article.
References
-
- Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BD, Burston HE, Vizeacoumar FJ, Snider J, Phanse S, et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. - PubMed
-
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J Royal Stat Soc Series B. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

