Tristetraprolin Limits Inflammatory Cytokine Production in Tumor-Associated Macrophages in an mRNA Decay-Independent Manner
- PMID: 26183929
- PMCID: PMC4526390
- DOI: 10.1158/0008-5472.CAN-15-0205
Tristetraprolin Limits Inflammatory Cytokine Production in Tumor-Associated Macrophages in an mRNA Decay-Independent Manner
Abstract
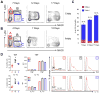
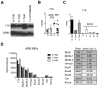
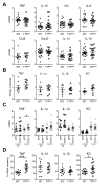
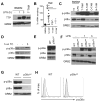
Tristetraprolin (TTP) is an inducible zinc finger AU-rich RNA-binding protein essential for enforcing degradation of mRNAs encoding inflammatory chemokines and cytokines. Most studies on TTP center on the connection between mRNA half-life and inflammatory output, because loss of TTP amplifies inflammation by increasing the stability of AU-rich mRNAs. Here, we focused on how TTP controls cytokine and chemokine production in the nonresolving inflammation of cancer using tissue-specific approaches. In contrast with model in vitro macrophage systems, we found constitutive TTP expression in late-stage tumor-associated macrophages (TAM). However, TTP's effects on AU-rich mRNA stability were negligible and limited by constitutive p38α MAPK activity, which was the main driver of proinflammatory cytokine production in TAMs at the posttranscriptional level. Instead, elimination of TTP caused excessive protein production of inflammatory mediators, suggesting TTP-dependent translational suppression of AU-rich mRNAs. Manipulation of the p38α-TTP axis in macrophages has significant effects on the growth of tumors and therefore represents a means to manipulate inflammation in the tumor microenvironment.
©2015 American Association for Cancer Research.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
The p38 MAPK pathway inhibits tristetraprolin-directed decay of interleukin-10 and pro-inflammatory mediator mRNAs in murine macrophages.FEBS Lett. 2009 Jun 18;583(12):1933-8. doi: 10.1016/j.febslet.2009.04.039. Epub 2009 May 3. FEBS Lett. 2009. PMID: 19416727 Free PMC article.
-
A Knock-In Tristetraprolin (TTP) Zinc Finger Point Mutation in Mice: Comparison with Complete TTP Deficiency.Mol Cell Biol. 2018 Jan 29;38(4):e00488-17. doi: 10.1128/MCB.00488-17. Print 2018 Feb 15. Mol Cell Biol. 2018. PMID: 29203639 Free PMC article.
-
Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation.Mol Syst Biol. 2011 Dec 20;7:560. doi: 10.1038/msb.2011.93. Mol Syst Biol. 2011. PMID: 22186734 Free PMC article.
-
Control of mRNA decay by phosphorylation of tristetraprolin.Biochem Soc Trans. 2008 Jun;36(Pt 3):491-6. doi: 10.1042/BST0360491. Biochem Soc Trans. 2008. PMID: 18481987 Review.
-
The role of tristetraprolin in cancer and inflammation.Front Biosci (Landmark Ed). 2012 Jan 1;17(1):174-88. doi: 10.2741/3920. Front Biosci (Landmark Ed). 2012. PMID: 22201737 Free PMC article. Review.
Cited by
-
Improving Dietary Intake of Essential Nutrients Can Ameliorate Inflammation in Patients with Diabetic Foot Ulcers.Nutrients. 2022 Jun 9;14(12):2393. doi: 10.3390/nu14122393. Nutrients. 2022. PMID: 35745123 Free PMC article. Clinical Trial.
-
Myeloid-specific deletion of Zfp36 protects against insulin resistance and fatty liver in diet-induced obese mice.Am J Physiol Endocrinol Metab. 2018 Oct 1;315(4):E676-E693. doi: 10.1152/ajpendo.00224.2017. Epub 2018 Mar 6. Am J Physiol Endocrinol Metab. 2018. PMID: 29509432 Free PMC article.
-
Analysis of early mesothelial cell responses to Staphylococcus epidermidis isolated from patients with peritoneal dialysis-associated peritonitis.PLoS One. 2017 May 24;12(5):e0178151. doi: 10.1371/journal.pone.0178151. eCollection 2017. PLoS One. 2017. PMID: 28542390 Free PMC article.
-
The Tristetraprolin Family of RNA-Binding Proteins in Cancer: Progress and Future Prospects.Cancers (Basel). 2020 Jun 11;12(6):1539. doi: 10.3390/cancers12061539. Cancers (Basel). 2020. PMID: 32545247 Free PMC article. Review.
-
Analysis of the Trichuris suis excretory/secretory proteins as a function of life cycle stage and their immunomodulatory properties.Sci Rep. 2018 Oct 29;8(1):15921. doi: 10.1038/s41598-018-34174-4. Sci Rep. 2018. PMID: 30374177 Free PMC article.
References
-
- Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140(6):871–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

