Biofilm Formation As a Response to Ecological Competition
- PMID: 26158271
- PMCID: PMC4497666
- DOI: 10.1371/journal.pbio.1002191
Biofilm Formation As a Response to Ecological Competition
Erratum in
-
Correction: Biofilm Formation As a Response to Ecological Competition.PLoS Biol. 2015 Aug 12;13(8):e1002232. doi: 10.1371/journal.pbio.1002232. eCollection 2015 Aug. PLoS Biol. 2015. PMID: 26266815 Free PMC article. No abstract available.
Abstract
Bacteria form dense surface-associated communities known as biofilms that are central to their persistence and how they affect us. Biofilm formation is commonly viewed as a cooperative enterprise, where strains and species work together for a common goal. Here we explore an alternative model: biofilm formation is a response to ecological competition. We co-cultured a diverse collection of natural isolates of the opportunistic pathogen Pseudomonas aeruginosa and studied the effect on biofilm formation. We show that strain mixing reliably increases biofilm formation compared to unmixed conditions. Importantly, strain mixing leads to strong competition: one strain dominates and largely excludes the other from the biofilm. Furthermore, we show that pyocins, narrow-spectrum antibiotics made by other P. aeruginosa strains, can stimulate biofilm formation by increasing the attachment of cells. Side-by-side comparisons using microfluidic assays suggest that the increase in biofilm occurs due to a general response to cellular damage: a comparable biofilm response occurs for pyocins that disrupt membranes as for commercial antibiotics that damage DNA, inhibit protein synthesis or transcription. Our data show that bacteria increase biofilm formation in response to ecological competition that is detected by antibiotic stress. This is inconsistent with the idea that sub-lethal concentrations of antibiotics are cooperative signals that coordinate microbial communities, as is often concluded. Instead, our work is consistent with competition sensing where low-levels of antibiotics are used to detect and respond to the competing genotypes that produce them.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
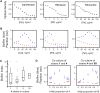

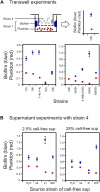
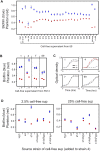
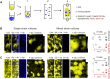

Comment in
-
Biofilms: How Structure Emerges from Conflict.Curr Biol. 2015 Sep 21;25(18):R800-2. doi: 10.1016/j.cub.2015.07.007. Curr Biol. 2015. PMID: 26394102
Similar articles
-
Competition in Biofilms between Cystic Fibrosis Isolates of Pseudomonas aeruginosa Is Shaped by R-Pyocins.mBio. 2019 Jan 29;10(1):e01828-18. doi: 10.1128/mBio.01828-18. mBio. 2019. PMID: 30696740 Free PMC article.
-
Biofilms: How Structure Emerges from Conflict.Curr Biol. 2015 Sep 21;25(18):R800-2. doi: 10.1016/j.cub.2015.07.007. Curr Biol. 2015. PMID: 26394102
-
Plant-expressed pyocins for control of Pseudomonas aeruginosa.PLoS One. 2017 Oct 3;12(10):e0185782. doi: 10.1371/journal.pone.0185782. eCollection 2017. PLoS One. 2017. PMID: 28973027 Free PMC article.
-
An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal.FEMS Immunol Med Microbiol. 2010 Aug;59(3):253-68. doi: 10.1111/j.1574-695X.2010.00690.x. Epub 2010 Apr 23. FEMS Immunol Med Microbiol. 2010. PMID: 20497222 Review.
-
Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies.J Biotechnol. 2014 Dec 10;191:121-30. doi: 10.1016/j.jbiotec.2014.09.003. Epub 2014 Sep 18. J Biotechnol. 2014. PMID: 25240440 Review.
Cited by
-
Overshadow Effect of Psl on Bacterial Response to Physiochemically Distinct Surfaces Through Motility-Based Characterization.Front Cell Infect Microbiol. 2018 Oct 29;8:383. doi: 10.3389/fcimb.2018.00383. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30420944 Free PMC article.
-
Biofilm through the Looking Glass: A Microbial Food Safety Perspective.Pathogens. 2022 Mar 12;11(3):346. doi: 10.3390/pathogens11030346. Pathogens. 2022. PMID: 35335670 Free PMC article. Review.
-
Geometrical Distribution of Cryptococcus neoformans Mediates Flower-Like Biofilm Development.Front Microbiol. 2017 Dec 19;8:2534. doi: 10.3389/fmicb.2017.02534. eCollection 2017. Front Microbiol. 2017. PMID: 29312225 Free PMC article.
-
Formation of a Mixed-Species Biofilm Is a Survival Strategy for Unculturable Lactic Acid Bacteria and Saccharomyces cerevisiae in Daqu, a Chinese Traditional Fermentation Starter.Front Microbiol. 2020 Feb 6;11:138. doi: 10.3389/fmicb.2020.00138. eCollection 2020. Front Microbiol. 2020. PMID: 32117157 Free PMC article.
-
Antibiotic Stimulation of a Bacillus subtilis Migratory Response.mSphere. 2018 Feb 21;3(1):e00586-17. doi: 10.1128/mSphere.00586-17. eCollection 2018 Jan-Feb. mSphere. 2018. PMID: 29507890 Free PMC article.
References
-
- Costerton JW (1995) Overview of microbial biofilms. J Ind Microbiol 15: 137–140. - PubMed
-
- Kolter R, Greenberg EP (2006) Microbial sciences—The superficial life of microbes. Nature 441: 300–302. - PubMed
-
- Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: From the natural environment to infectious diseases. Nature Reviews Microbiology 2: 95–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

